Following on from (but unrelated to) my post last week about feed tools we have two posts, one from Deepak Singh, and one from Neil Saunders, both talking about ‘friend feeds’ or ‘lifestreams’. The idea here is of aggregating all the content you are generating (or is being generated about you?) into one place. There are a couple of these about but the main ones seem to be Friendfeed and Profiliac. See Deepaks’s post (or indeed his Friendfeed) for details of the conversations that can come out of these type of things.
What piqued my interest though was the comment Neil made at the bottom of his post about Workstreams.
Here’s a crazy idea – the workstream:
* Neil parsed SwissProt entry Q38897 using parser script swiss2features.pl
* Bob calculated all intersubunit contacts in PDB entry 2jdq using CCP4 package contact
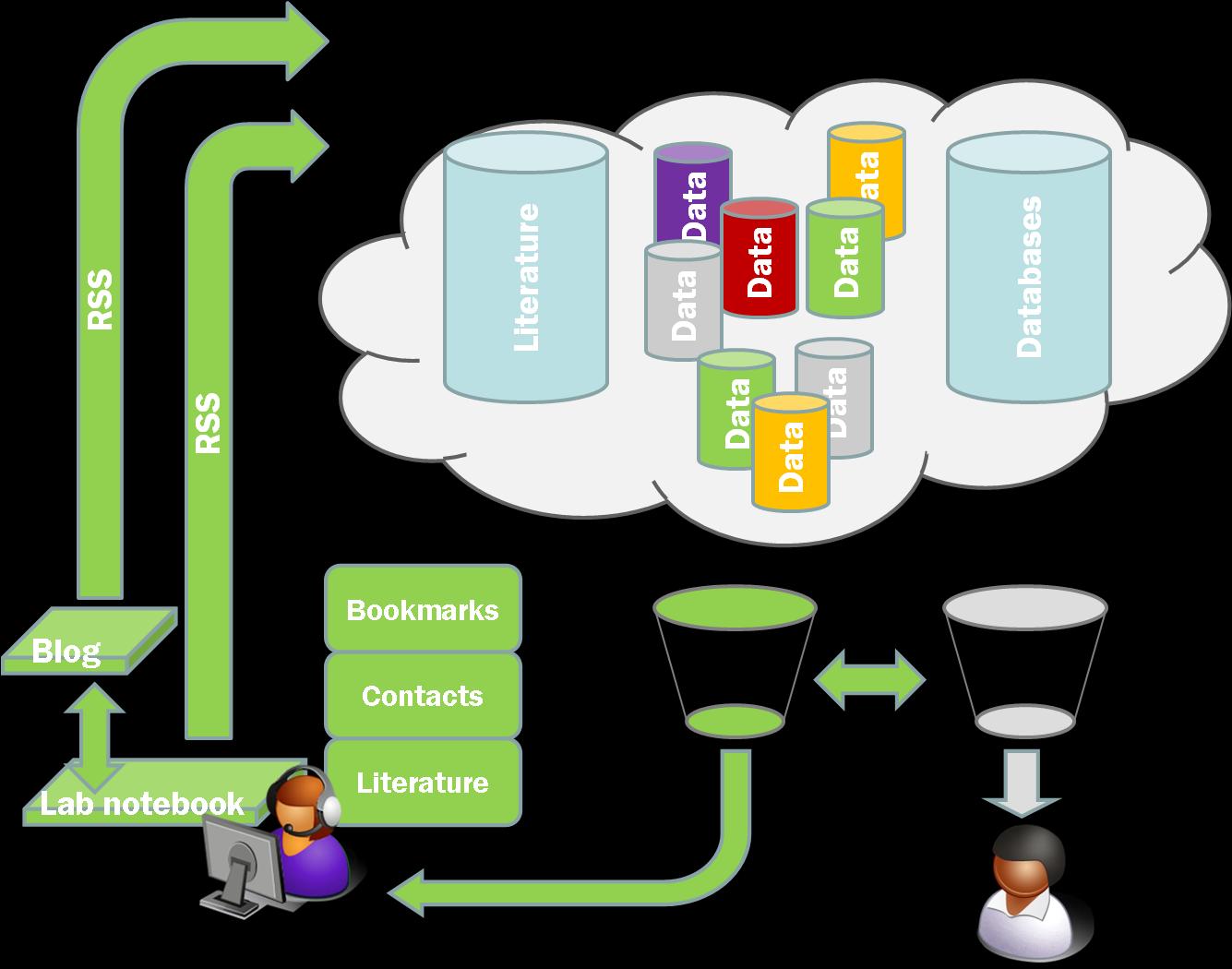
This is exactly the kind of thing I was thinking about as the raw material for the aggregators that would suggest things that you ought to look at, whether it be a paper, a blog post, a person, or a specific experimental result. This type of system will rely absolutely on the willingness of people to make public what they are reading, doing, even perhaps thinking. Indeed I think this is the raw information that will make another one of Neil’s great suggestions feasible.
Following on from Neil’s post I had a short conversation with Alf in the comments about blogging (or Twittering) machines. Alf pointed out a really quite cool example. This is something that we are close to implementing in the open in the lab at RAL. We hope to have the autoclave, PCR machine, and balances all blogging out what they are seeing. This will generate a data feed that we can use to pull specific data items down into the LaBLog.
Perhaps more interesting is the idea of connecting this to people. At the moment the model is that the instruments are doing the blogging. This is probably a good way to go because it keeps a straightforward identifiable data stream. At the moment the trigger for the instruments to blog is a button. However at RAL we use RFID proximity cards for access to the buildings. This means we have an easy identifier for people, so what we aim to do is use the RFID card to trigger data collection (or data feeding).
If this could be captured and processed there is the potential for capturing a lot of the detail of what has happened in the laboratory. Combine this with a couple of Twitter posts giving a little more personal context and it may be possible to reconstruct a pretty complete record of what was done and precisely when. The primary benefit of this would be in trouble shooting but if we could get a little bit of processing into this, and if there are specific actions with agreed labels, then it may be possible to automatically create a large portion of the lab book record.
This may be a great way of recording the kind of machine readable description of experiments that Jean-Claude has been posting about. Imagine a simplistic Twitter interface where you have a limited set of options (I am stirring, I am mixing, I am vortexing, I have run a TLC, I have added some compound). Combine this with a balance, a scanner, and a heating mantle which are blogging out what they are currently seeing, and a barcode reader (and printer) so as to identify what is being manipulated and which compound is which.
One of the problems we have with our lab books is that they can never be detailed enough to capture everything that somebody might be interested in one day. However at the same time they are too detailed for easy reading by third parties. I think there is general agreement that on top of the lab book you need an interpretation layer, an extra blog that explains what is going on to the general public. Perhaps by capturing all the detailed bits automatically we can focus on planning and thinking about the experiments rather than worrying about how to capture everything manually. Then anyone can mash up the results, or the discussion, or the average speed of the stirrer bar, any way they like.