ChemSpidey lives! Even in the face of Karen James’ heavy irony I am still amazed that someone like me with very little programming experience was able to pull together something that actually worked effectively in a live demo. As long as you’re not actively scared of trying to put things together it is becoming relatively straightforward to build tools that do useful things. Building ChemSpidey relied heavily on existing services and other people’s code but pulling that together was a relatively straightforward process. The biggest problems were fixing the strange and in most cases undocumented behaviour of some of the pieces I used. So what is ChemSpidey?
ChemSpidey is a Wave robot that can be found at chemspidey@appspot.com. The code repository is available at Github and you should feel free to re-use it in anyway you see fit, although I wouldn’t really recommend it at the moment, it isn’t exactly the highest quality. One of the first applications I see for Wave is to make it easy to author (semi-)semantic documents which link objects within the document to records on the web. In chemistry it would be helpful to link the names of compounds through to records about those compounds on the relevant databases.
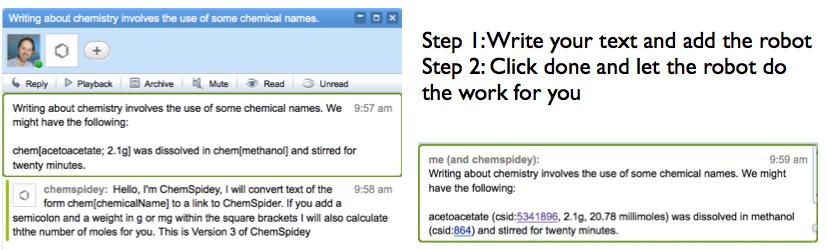
If ChemSpidey is added to a wave it watches for text of the form “chem[ChemicalName{;weight {m}g}]” where the curly bracketed parts are optional. When a blip is submitted by hitting the “done” button ChemSpidey searches through the blip looking for this text and if it finds it, strips out the name and sends it) to the ChemSpider SimpleSearch service. ChemSpider returns a list of database ids and the robot currently just pulls the top one off the list and adds the text ChemicalName (csid:####) to the wave, where the id is linked back to ChemSpider. If there is a weight present it asks the ChemSpider MassSpec API for the nominal molecular weight calculates the number of moles and inserts that. You can see video of it working here (look along the timeline for the ChemSpidey tag).
What have I learned? Well some stuff that is probably obvious to anyone who is a proper developer. Use the current version of the API. Google AppEngine pushes strings around as unicode which broke my tests because I had developed things using standard Python strings. But I think it might be useful to start drawing some more general lessons about how best to design robots for research, so to kick of the discussion here are my thoughts, many of which came out of discussions with Ian Mulvany as we prepared for last weeks demo.
- Always add a Welcome Blip when the Robot is added to a wave. This makes the user confident that something has happened. Lets you notify users if a new version has been released, which might change the way the robot works, and lets you provide some short instructions.It’s good to include a version number here as well.
- Have some help available. Ian’s Janey robot responds to the request (janey help) in a blip with an extended help blip explaining context. Blips are easily deleted later if the user wants to get rid of them and putting these in separate blips keeps them out of the main document.
- Where you modify text leave an annotation. I’ve only just started to play with annotations but it seems immensely useful to at least attempt to leave a trace here of what you’ve done that makes it easy for either your own Robot or others, or just human users to see who did what. I would suggest leaving annotations that identfy the robot, include any text that was parsed, and ideally provide some domain information. We need to discuss how to setup some name spaces for this.
- Try to isolate the “science handling” from the “wave handling”. ChemSpidey mixes up a lot of things into one Python script. Looking back at it now it makes much more sense to isolate the interaction with the wave from the routines that parse text, or do mole calculations. This means both that the different levels of code should become easier for others to re-use and also if Wave doesn’t turn out to be the one system to rule them all that we can re-use the code. I am no architecture expert and it would be good to get some clues from some good ones about how best to separate things out.
These are just some initial thoughts from a very novice Python programmer. My code satisfies essentially none of these suggestions but I will make a concerted attempt to improve on that. What I really want to do is kick the conversation on from where we are at the moment, which is basically playing around, into how we design an architecture that allows rapid development of useful and powerful functionality.
