- Image via Wikipedia
I am frequently overly enamoured of the idea of where we might get to, forgetting that there are a lot of people still getting used to where we’ve been. I was forcibly reminded of this by Carole Goble on the weekend when I expressed a dislike of the Utopia PDF viewer that enables active figures and semantic markup of the PDFs of scientific papers. “Why can’t we just do this on the web?” I asked, and Carole pointed out the obvious, most people don’t read papers on the web. We know it’s a functionally better and simpler way to do it, but that improvement in functionality and simplicity is not immediately clear to, or in many cases even useable by, someone who is more comfortable with the printed page.
In my defence I never got to make the second part of the argument which is that with the new generation of tablet devices, lead by the iPad, there is a tremendous potential to build active, dynamic and (under the hood hidden from the user) semantically backed representations of papers that are both beautiful and functional. The technical means, and the design basis to suck people into web-based representations of research are falling into place and this is tremendously exciting.
However while the triumph of the iPad in the medium term may seem assured, my record on predicting the impact of technical innovations is not so good given the decision by Google to pull out of futher development of Wave primarily due to lack of uptake. Given that I was amongst the most bullish and positive of Wave advocates and yet I hadn’t managed to get onto the main site for perhaps a month or so, this cannot be terribly surprising but it is disappointing.
The reasons for lack of adoption have been well rehearsed in many places (see the Wikipedia page or Google News for criticisms). The interface was confusing, a lack of clarity as to what Wave is for, and simply the amount of user contribution required to build something useful. Nonetheless Wave remains for me an extremely exciting view of the possibilites. Above all it was the ability for users or communities to build dynamic functionality into documents and to make this part of the fabric of the web that was important to me. Indeed one of the most important criticisms for me was PT Sefton’s complaint that Wave didn’t leverage HTML formatting, that it was in a sense not a proper part of the document web ecosystem.
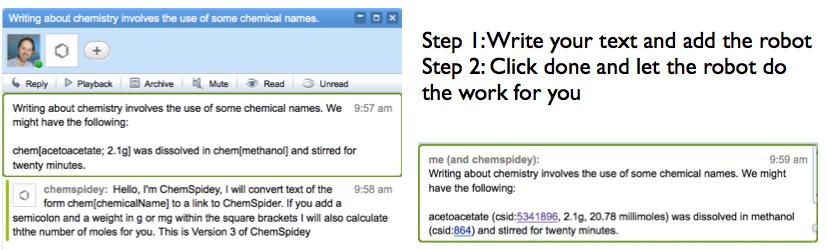
The key for me about the promise of Wave was its ability to interact with web based functionality, to be dynamic; fundamentally to treat a growing document as data and present that data in new and interesting ways. In the end this was probably just too abstruse a concept to grab hold of a user. While single demonstrations were easy to put together, building graphs, showing chemistry, marking up text, it was the bigger picture that this was generally possible that never made it through.
I think this is part of the bigger problem, similar to that we experience with trying to break people out of the PDF habit that we are conceptually stuck in a world of communicating through static documents. There is an almost obsessive need to control the layout and look of documents. This can become hilarious when you see TeX users complaining about having to use Word and Word users complaining about having to use TeX for fundamentally the same reason, that they feel a loss of control over the layout of their document. Documents that move, resize, or respond really seem to put people off. I notice this myself with badly laid out pages with dynamic sidebars that shift around, inducing a strange form of motion sickness.
There seems to be a higher aesthetic bar that needs to be reached for dynamic content, something that has been rarely achieved on the web until recently and virtually never in the presentation of scientific papers. While I philosophically disagree with Apple’s iron grip over their presentation ecosystem I have to admit that this has made it easier, if not quite yet automatic, to build beautiful, functional, and dynamic interfaces.
The rapid development of tablets that we can expect, as the rough and ready, but more flexible and open platforms do battle with the closed but elegant and safe environment provided by the iPad, offer real possibilities that we can overcome this psychological hurdle. Does this mean that we might finally see the end of the hegemony of the static document, that we can finally consign the PDF to the dustbin of temporary fixes where it belongs? I’m not sure I want to stick my neck out quite so far again, quite so soon and say that this will happen, or offer a timeline. But I hope it does, and I hope it does soon.
Related articles by Zemanta
- Google Bails on Wave (wired.com)
- So Long, Wave, But You’ll Live On In Your Successors (webworkerdaily.com)
- Google Wave is dead! Long live Google Wave! (programmableweb.com)
- Google’s Departing Wave (cyberculturalist.com)
- Apathy kills Google’s new-age Wave (go.theregister.com)