“In the future everyone will be a journal editor for 15 minutes” – apologies to Andy Warhol
Suddenly it seems everyone wants to re-imagine scientific communication. From the ACS symposium a few weeks back to a PLoS Forum, via interesting conversations with a range of publishers, funders and scientists, it seems a lot of people are thinking much more seriously about how to make scientific communication more effective, more appropriate to the 21st century and above all, to take more advantage of the power of the web.
For me, the “paper” of the future has to encompass much more than just the narrative descriptions of processed results we have today. It needs to support a much more diverse range of publication types, data, software, processes, protocols, and ideas, as well provide a rich and interactive means of diving into the detail where the user in interested and skimming over the surface where they are not. It needs to provide re-visualisation and streaming under the users control and crucially it needs to provide the ability to repackage the content for new purposes; education, public engagement, even main stream media reporting.
I’ve got a lot of mileage recently out of thinking about how to organise data and records by ignoring the actual formats and thinking more about what the objects I’m dealing with are, what they represent, and what I want to do with them. So what do we get if we apply this thinking to the scholarly published article?
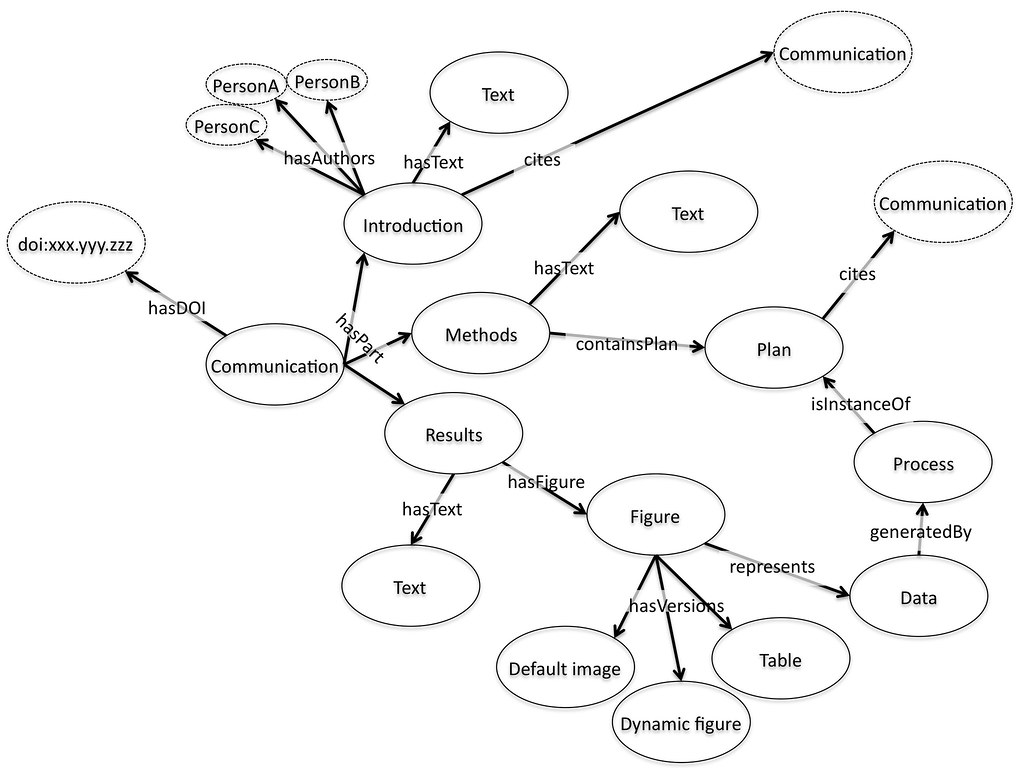
For me, a paper is an aggregation of objects. It contains, text, divided up into sections, often with references to other pieces of work. Some of these references are internal, to figures and tables, which are representations of data in some form or another. The paper world of journals has led us to think about these as images but a much better mental model for figures on the web is of an embedded object, perhaps a visualisation from a service like Many Eyes, Swivel, and Tableau Public. Why is this better? It is better because it maps more effectively onto what we want to do with the figure. We want to use it to absorb the data it represents, and to do this we might want to zoom, pan, re-colour, or re-draw the data. But we want to know if we do this that we are using the same underlying data, so the data needs a home, an address somewhere on the web, perhaps with the journal, or perhaps somewhere else entirely, that we can refer to with confidence.
If that data has an individual identity it can in turn refer back to the process used to generate it, perhaps in an online notebook or record, perhaps pointing to a workflow or software process based on another website. Maybe when I read the paper I want that included, maybe when you read it you don’t – it is a personal choice, but one that should be easy to make. Indeed, it is a choice that would be easy to make with today’s flexible web frameworks if the underlying pieces were available and represented in the right way.
The authors of the paper can also be included as a reference to a unique identifier. Perhaps the authors of the different segments are different. This is no problem, each piece can refer to the people that generated it. Funders and other supporting players might be included by reference. Again this solves a real problem of today, different players are interested in how people contributed to a piece of work, not just who wrote the paper. Providing a reference to a person where the link show what their contribution was can provide this much more detailed information. Finally the overall aggregation of pieces that is brought together and finally published also has a unique identifier, often in the form of the familiar DOI.
This view of the paper is interesting to me for two reasons. The first is that it natively supports a wide range of publication or communication types, including data papers, process papers, protocols, ideas and proposals. If we think of publication as the act of bringing a set of things together and providing them with a coherent identity then that publication can be many things with many possible uses. In a sense this is doing what a traditional paper should do, bringing all the relevant information into a single set of pages that can be found together, as opposed to what they usually do, tick a set of boxes about what a paper is supposed to look like. “Is this publishable?” is an almost meaningless question on the web. Of course it is. “Is it a paper?” is the question we are actually asking. By applying the principles of what the paper should be doing as opposed to the straightjacket of a paginated, print-based document, we get much more flexibility.
The second aspect which I find exciting revolves around the idea of citation as both internal and external references about the relationships between these individual objects. If the whole aggregation has an address on the web via a doi or a URL, and if its relationship both to the objects that make it up and to other available things on the web are made clear in a machine readable citation then we have the beginnings of a machine readable scientific web of knowledge. If we take this view of objects and aggregates that cite each other, and we provide details of what the citations mean (this was used in that, this process created that output, this paper is cited as an input to that one) then we are building the semantic web as a byproduct of what we want to do anyway. Instead of scaring people with angle brackets we are using a paradigm that researchers understand and respect, citation, to build up meaningful links between packages of knowledge. We need the authoring tools that help us build and aggregate these objects together and tools that make forming these citations easy and natural by using the existing ideas around linking and referencing but if we can build those we get the semantic web for science as a free side product – while also making it easier for humans to find the details they’re looking for.
Finally this view blows apart the monolithic role of the publisher and creates an implicit marketplace where anybody can offer aggregations that they have created to potential customers. This might range from a high school student putting their science library project on the web through to a large scale commercial publisher that provides a strong brand identity, quality filtering, and added value through their infrastructure or services. And everything in between. It would mean that large scale publishers would have to compete directly with the small scale on a value-for-money basis and that new types of communication could be rapidly prototyped and deployed.
There are a whole series of technical questions wrapped up in this view, in particular if we are aggregating things that are on the web, how did they get there in the first place, and what authoring tools will we need to pull them together. I’ll try to start on that in a follow-up post.

![Reblog this post [with Zemanta]](http://img.zemanta.com/reblog_e.png?x-id=8af89e7c-59ca-438d-8e68-4efa40cffd79)
This makes sense to me. That's a pretty good starting point as far as I am concerned. :)
I'm at a PLoS workshop on “biodiversity hubs” at the California where there's been a spectacular lack of clarity about what a hub should actually be (certainly PLoS haven't articulated it clearly). The vision you outline here is cool, and goes beyond simply expanding on how we display scientific articles, which is where the publishing industry seems to be heading. In my own efforts to re-imagine the scientific article (http://hdl.handle.net/10101/npre.2009.3173.1 and http://iphylo.org/~rpage/challenge) I also disassembled the article into pieces (primarily authors, sequences, specimens, and taxa), and offered views where those objects (not the article) are the primary focus, but I didn't explore at ways these could be recombined, which is where you're heading, and where the real fun starts. I guess we are still overly focussed on articles as the fundamental unit, which is what we need to overcome if we are to make real progress, which is why I've been somewhat underwhelmed by semantic tagging of articles as an end in themselves http://iphylo.blogspot.com/2009/04/semantic-pub…
The approach you outline is likely to work best in disciplines that are data rich, where data objects have a digital presence (and identifier), and the data have a lifetime beyond that of an individual paper (e.g., can be reused in different contexts). Looking forward to the follow up post…
There are many of us that share this view. However, having been working as a web developer for some time I can tell you there's a little change that I'd make to it. There shouldn't be any “unique identifiers”. It'd be cool if they existed, but they're quite incompatible with how the web works. What's needed in order to start working on taking the next step are web standards for exposing all this data.
Your definitional statement of “what is a paper?” is excellent. In such an online publishing environment I wonder how peer review would evolve. I can imagine it being much more dynamic, though qualified reviewers would need some kind of cross-site authority ranking.
In any case, thanks for the thoughts. We created Tableau Public to support scenarios such as online publishing and commenting and would love to see it used that way.
Great post. this was in part foreshadowed, ” Blurring the boundaries between the scientific 'papers' and biological databases”: http://www.nature.com/nature/debates/e-access/A…
A related issue is how to highlight papers within a given field? Ovelay jnls such as AIP's Virtual Journals series have aggregated content from a number of societies and publishers such as Nature. Faculty of 1000 reviews and ranks papers, but it is a subscription service. Elsevier now features article tools that link to citations in Scopus and biological information in NextBio. But how best to serve the researcher who has to make a choice between running experiments and reading the literature and wants to stay informed about the key papers in her/his field?
Great post. this was in part foreshadowed, ” Blurring the boundaries between the scientific 'papers' and biological databases”: http://www.nature.com/nature/debates/e-access/A…
A related issue is how to highlight papers within a given field? Ovelay jnls such as AIP's Virtual Journals series have aggregated content from a number of societies and publishers such as Nature. Faculty of 1000 reviews and ranks papers, but it is a subscription service. Elsevier now features article tools that link to citations in Scopus and biological information in NextBio. But how best to serve the researcher who has to make a choice between running experiments and reading the literature and wants to stay informed about the key papers in her/his field?
Hi Garrett, my views on highlighting papers within a field is that this is probably the wrong approach. I would argue for highlighting papers for the specific user using a mixture of search (probably smarter and more semantic than today's default) but crucially much more social information about other people interacting with the same objects. My personal view is that disciplines and communities are so fragments and dynamics that statically serving them is not helpful.
That said there is a case for using overlay journals as community sites – ways of sharing and interacting around the papers for a community. But I think this may be a step towards something much more dynamic, a personal overlay journal that changes as you and your research social network change. We are a long way away from that though.
Peer review and how it relates to aggregated objects is an interesting question. I've got pretty heretical views on peer review anyway so I'm probably not the person to ask, but I can see much more value being gained from knowing who has commented on what aspect of a particular portion of a paper. I may have much more respect e.g. for your assessment of the statistical validity of the conclusions than your assessment of the overall importance of the finding, or vice versa. But this depends on an open and federated peer review system as well and we are some way away from that!
A question that was raised offline was the one of business models for data visualisation services. Do you have any feel for how this is playing out? Who would you see as paying for the visualisation that appears on a third party website. Does the publisher pay for the functionality and pass that on to the author, does the author pay in the preparation of their presentation, or does the end user pay for access to the visualisation?
Juan, I think we might be at cross purposes but correct me if I'm wrong. By a unique identifier in this context I generally mean a URL. We need to be able to find things one way or another so that address for me is the unique identifier.
An example of an author tool is Escape, developed as a SURF project (The Netherlands) carried out by University of Groningen, University of Twente and the Royal Academy of Arts and Sciences. See: http://escapesurf.wordpress.com/. It is still a proof of concept, just look at for instance: http://escape.utwente.nl/show/191?graph=1 and click around. Although many objects are in Dutch it gives you a view of how the aggregation is structured.
Visualization that appears on a third-part site is usually paid for by the site. In the case of Tableau Public, which is a free tool, no one pays. Our business (I work for Tableau) is selling visualization to companies, and we made Tableau Public free both to help awareness for our corporate business and because we care about good data on the public web.
In any case, It's in the interest of the website to pay because visualization can increase reader engagement and dwell time, which if the website wishes can be monetized through ad sales.
If the scientific article (or most of digital content for that matter) is -or becoming- an aggregate object, what are the implications for the information overload? On a Harvard blog devoted to the changing nature of information, we reference to your article and a new stream of ideas about new forms, causes and remedies for information overload. If -as David Weinberger pointed out a while ago- in the digital world everything is miscallenuous, and now the same process of disaggregation is taking place in scholarly publications, are we in fact ensuring the coming of a “cosmic scale” of information overload? We are already drowning in articles, now we will drown in article bits and pieces, and information aggregation will certainly become the future of communication. But is this a desired outcome?
I would love to invite you to write a reply to this question on our blog at globalknowledgexchange.net. We are attempting to understand the changing nature of information across all organizational types (not just in academia), and view information using the classis distinction made by Buckland (as-a-thing, as-a-process, as-knowledge).
Hi, and thanks for the thoughtful comment and the link in. I'll give the brief technophilic answer to your question as a starter. One of the reasons we are drowning in articles is because they don't link properly into the wider web and therefore don't enable us to use the set of tools (search, aggregation, indexing) that we are familiar with on the web. Search and discovery is poor for academic articles precisely because they are monolothic single text documents with a couple of images in them. By pulling that apart and providing lots of internal links that provide meaning we should be able to improve search and discovery. Yes there will be more stuff but there is always more stuff. And search is just getting better and better.
Cameron, is there a “discussion” section in your diagram? It seems that this bit of text should also be off the communication hub pointing to intro/method/results, with arrows between its text and results, and outward pointing arrows to other points of communication which themselves lead to other DOI/pubs (since the discussion is essentially a dialogue of support, contention, and rebuttal between sets of results). Would be really cool to model this in 3D, for one paper (probably a short one!).
Hi Mickey
Its a good question. I think discussion is actually implicit in the diagram.
The overall discussion is the set of all other communications that cite some
portion of the original. You can then imagine reducing that large set by
specifying all communications that come from some subset of sources
(friendfeed and twitter, or formally published reviews), or that cite just
the conclusion (or just the data, or…). By following the citation trail
you can also actually get a threaded discussion (or a webbed one if there
are cross citations).
An editor might aggregate some subset of these together to create a new
communication which would be a curated discussion based on some criteria as
well. But all in all this kind of view does some nice things like explicitly
connect formally published and informally published objects together.
As a reader of science, I like the idea of a “curated discussion” but I also
want space explicitly provided where the researchers themselves have the
opportunity to say what they think their research accomplished or how it
fits into the bigger picture. As writers of science, my students are often
dismayed when they get to the Discussion as a writing object b/c it is much
more limited in scope than they were hoping — I think this idea of a
dynamic paper addresses this problem nicely b/c it “detaches” each part of
the paper, meaning that a discussion can exist with criteria attached AND
there is still room for the looser, more interesting speculation that any
piece of research also provokes (the stuff I tell my students happens in
hallways and at conferences, especially at the bar). Also, just want to say
that I really like the diagram — it's taken most of the last year for me to
finally “get” this approach. Would love to see the figure animated, a la
the kind of stuff Thinkmap puts together for the visual thesaurus and other
data-heavy projects (http://www.thinkmap.com).
Sorry, I hadn't twigged what you were talking about. The “discussion section” of a traditional paper isn't there simply because I forgot to put it in! Yes, I take your point that this view enables a much more flexible way of imagining this (and indeed the other sections – methods might be an aggregation of code and scripts, or a makefile). Yep a dynamic diagram would be cool – a bit beyond my abilities at the moment though…
This is so wrong. Thinking of a paper as simply a collection of information is like thinking of a poem as simply a collection of words. Working within the constraints, of metric, rhyme and so on, struggling with the constraints, you often end up with a more beautiful poem. The same is true of any kind of authorship, with poetry just an extreme example.
Actually, it holds even more broadly. Compare developing on an iPhone to developing for a Blackberry, for example. Apply loosely enforces a set of interface constraints, and this makes life easier for both the developers and the users.
I disagree on two counts here. Firstly I never meant to say a paper is
simply a collection of information. In fact I would argue that my intention
is to raise the status of the composition process to primacy so as to enable
more creativity.
Secondly the role of the artist is arguably in choosing the _right_ set of
constraints for a particular composition. Constraints can be powerful and
focussing or they can be unhelpful or dangerous. You assume that our current
straighjacket is the right one. I would argue it is more like playing tennis
with one hand tied behind your back. The majority of papers published today
are _less_ than the sum of their parts. Ideas, data, and imagery that are
damaged, made less clear by being tied into an old fashioned framework. You
are saying that all poetry should be sonnets. I am saying that at the moment
we have haiku being rammed in as the heroic couple because sonnets are the
only way someone can get published
But I think you're iPhone analogy is apt. The iPhone is a closed system that
focuses on producing slick user experiences that match their expectations.
What it is very bad at is supporting radical innovation because the whole
environment makes taking risks and exploring new ideas difficult. It
channels behaviour and thinking in defined ways. This is exactly the
opposite of what we want in an optimised scientific environment. In science
we need generative systems that support the process of innovation, while
efficiently identifying blind alleys and mistakes. Science needs the madness
of open source. Technology and marketing can benefit from slick interfaces
and the iPhone approach. That is engineering, not science – and they have
very different needs.