An update on the Workshop that I announced previously. We have a number of people confirmed to come down and I need to start firming up numbers. I will be emailing a few people over the weekend so sorry if you get this via more than one route. The plan of attack remains as follows:
Meet on evening of Sunday 31 August in Southampton, most likely at a bar/restaurant near the University to coordinate/organise the details of sessions.
Commence on Monday at ~9:30 and finish around 4:30pm (with the option of discussion going into the evening) with three or four sessions over the course of the day broadly divided into the areas of tools, social issues, and policy. We have people interested and expert in all of these areas coming so we should be able to to have a good discussion. The object is to keep it very informal but to keep the discussion productive. Numbers are likely to be around 15-20 people. For those not lucky enough to be in the area we will aim to record and stream the sessions, probably using a combination of dimdim, mogulus, and slideshare. Some of these may require you to be signed into our session so if you are interested drop me a line at the account below.
To register for the meeting please send me an email to my gmail account (cameronneylon). To avoid any potential confusion, even if you have emailed me in the past week or so about this please email again so that I have a comprehensive list in one place. I will get back to you with a request via PayPal for £15 to cover coffees and lunch for the day (so if you have a PayPal account you want to use please send the email from that address). If there is a problem with the cost please state so in your email and we will see what we can do. We can suggest options for accomodation but will ask you to sort it out for yourself.
I have set up a wiki to discuss the workshop which is currently completely open access. If I see spam or hacking problems I will close it down to members only (so it would be helpful if you could create an account) but hopefully it might last a few weeks in the open form. Please add your name and any relevant details you are happy to give out to the Attendees page and add any presentations or demos you would be interested in giving, or would be interested in hearing about, on the Programme suggestion page.

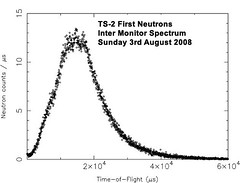 darkest Oxfordshire. I am based at
darkest Oxfordshire. I am based at 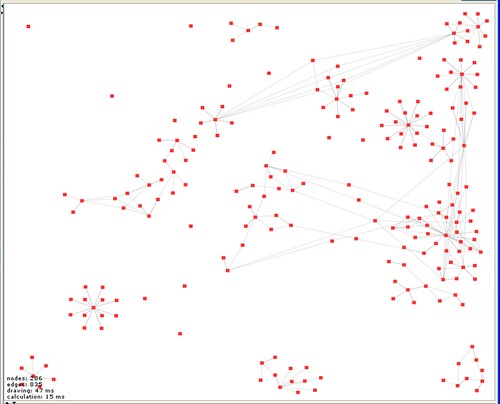 ings that are immediately obvious. The first is that I start a lot of things and don’t necessarily manage to get very far with them and that I do a number of (currently) unrelated things (isolated subgraphs aren’t connected). Also that there are some materials that get widely re-used and some that don’t. There are also clearly things that I haven’t finished entering properly (isolated nodes). Finally, that we need a more sophisticated tool for playing with the view because building a human readable version of the graph will require some manipulation, grabbing subgraphs and moving them around. Welkin is great but after 30 minutes playing I have a bunch of feature requests. But this is what I’ve done so far. I am sure there are many things that can be done with this kind of view – but for the moment what is important is that it is an entirely new kind of way of looking at the record.
ings that are immediately obvious. The first is that I start a lot of things and don’t necessarily manage to get very far with them and that I do a number of (currently) unrelated things (isolated subgraphs aren’t connected). Also that there are some materials that get widely re-used and some that don’t. There are also clearly things that I haven’t finished entering properly (isolated nodes). Finally, that we need a more sophisticated tool for playing with the view because building a human readable version of the graph will require some manipulation, grabbing subgraphs and moving them around. Welkin is great but after 30 minutes playing I have a bunch of feature requests. But this is what I’ve done so far. I am sure there are many things that can be done with this kind of view – but for the moment what is important is that it is an entirely new kind of way of looking at the record.