http://friendfeed.com/e/3865430a-3f03-4f58-9851-f8ba37d97f5b/Data-sharing-in-the-Biosciences-II/
Open Science Workshop at Southampton – 31 August and 1 September 2008
Image via Wikipedia
I’m aware I’ve been trailing this idea around for sometime now but its been difficult to pin down due to issues with room bookings. However I’m just going to go ahead and if we end up meeting in a local bar then so be it! If Southampton becomes too difficult I might organise to have it at RAL instead but Southampton is more convenient in many ways.
Science Blogging 2008: London will be held on August 30 at the Royal Institution and as a number of people are coming to that it seemed a good opportunity to get a few more people together to have a get together and discuss how we might move things forward. This now turns out to be one of a series of such workshops following on from Collaborating for the future of open science, organised by Science Commons as a satellite meeting of EuroScience Open Forum in Barcelona next month, BioBarCamp/Scifoo from 5-10 August and a possible Open Science Workshop at Stanford on Monday 11 August, as well as the Open Science Workshop in Hawaii (can’t let the bioinformaticians have all the good conference sites to themselves!) at the Pacific Symposium on Biocomputing.
For the Southampton meeting I would propose that we essentially look at having four themed sessions: Tools, Data standards, Policy/Funding, and Projects. Within this we adopt an unconference style where we decide who speaks based on who is there and want to present something. My ideas is essentially to meet on the Sunday evening at a local hostelry to discuss and organise the specifics of the program for Monday. On the Monday we spend the day with presentations and leave plenty of room for discussion. People can leave in the afternoon, or hang around into the evening for further discussion. We have absolutely zero, zilch, nada funding available so I will be asking for a contribution (to be finalised later but probably £10-15 each) to cover coffee/tea and lunch on the Monday.
Travel plans
Image via Wikipedia
For anyone who is interested I thought it might be helpful to say where I am going to be at meetings and conferences over the next few months. If anyone is going then drop me a line and we can meet up. Most of these are looking like interesting meetings so I recommend going to them if they are in your area (and in your area if you see what I mean) Continue reading “Travel plans”
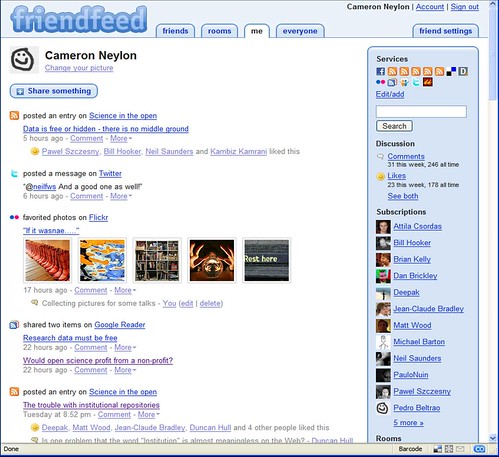
Friendfeed for scientists: What, why, and how?
There has been lots of interest amongst some parts of the community about what has been happening on FriendFeed. A growing number of people are signed up and lots of interesting conversations are happening. However it was suggested that as these groups grow they become harder to manage and the perceived barriers to entry get higher. So this is an attempt to provide a brief intro to FriendFeed for the scientist who may be interested in using it; what it is, why it is useful, and some suggestions on how to get involved without getting overwhelmed. This are entirely my views and your mileage may obviously vary.
What is FriendFeed?
FriendFeed is a ‘lifestreaming’ service or more simply a personal aggregator. It takes data streams that you generate and brings them all together into one place where people can see them. You choose to subscribe to any of the feeds you already generate (Flickr stream, blog posts, favorited YouTube videos, and lots of other services integrated). In addition you can post links to specific web pages or just comments into your stream. A number of these types of services have popped up in the recent months including Profilactic and Social Thing but FriendFeed has two key aspects that have led it to the fore. Firstly the commenting facilities enable rapid and effective conversations and secondly there was rapid adoption by a group of life scientists which has created a community. Like anything some of the other services have advantages and probably have their own communities but for science and in particular the life sciences FriendFeed is where it is at.
As well as allowing other people to look at what you have been doing FriendFeed allows you to subscribe to other people and see what they have been doing. You have the option of ‘liking’ particular items and commenting on them. In addition to seeing the items of your friends, people you are subscribed to, you also see items that they have liked or commented on. This helps you to find new people you may be interested in following. It also helps people to find you. As well as this items with comments or likes then get popped up to the top of the feed so items that are generating a conversation keep coming back to your attention.
These conversations can happen very fast. Some conversations baloon within minutes, most take place at a more sedate pace over a couple of hours or days but it is important to be aware that many people are live most of the time.
Why is FriendFeed useful?
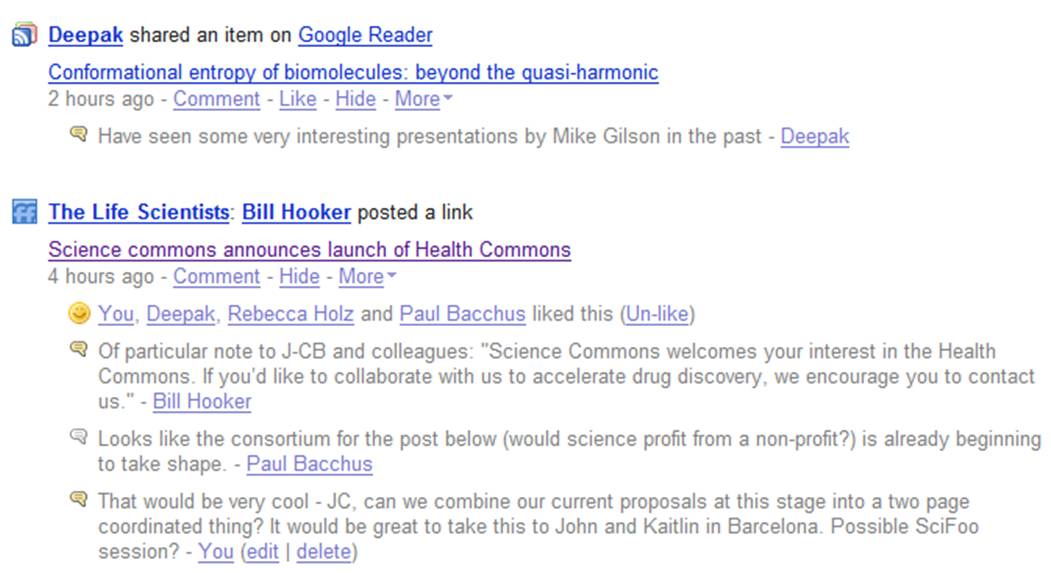
So how is FriendFeed useful to a scientist? First and foremost it is a great way of getting rapid notification of interesting content from people you trust. Obviously this depends on there people who are interested in the same kinds of things that you are but this is something that will grow as the community grows. A number of FriendFeed users stream both del.icio.us bookmark pages as well as papers or web articles they have put into citeulike or connotea or simply via sharing it in Google Reader. Also you can get information that people have shared on opportunities, meetings, or just interesting material on the web. Think of it as an informal but continually running journal club – always searching for the next thing you will need to know about.
But FriendFeed is about much more than finding things on the web. One of its most powerful features is the conversations that can take place. Queries can be answered very rapidly going some way towards making possible the rapid formation of collaborative networks that can come together to solve a specific problem. Its not there yet but there are a growing number of examples where specific ideas were encouraged, developed, or problems solved quickly by bringing the right expertise to bear.
One example is shown in the following figure where I was looking for some help in building a particular protein model for a proposal. I didn’t really know how to go about this and didn’t have the appropriate software to hand. Pawel Szczesny offered to help and was able to quickly come up with what I wanted. In the future we hope to generate data which Pawel may be able to help us analyse. You can see the whole story and how it unfolded after this at http://friendfeed.com/search?q=mthk
We are still a long way from the dream of just putting out a request and getting an answer but it is worth point out that the whole exchange here lasted about four hours. Other collaborative efforts have also formed, most recently leading to the formation of BioGang, a collaborative area for people to work up and comment on possible projects.
So how do I use it? Will I be able to cope?
FriendFeed can be as high volume as you want it be but if its going to be useful to you it has to be manageable. If you’re the kind of person who already manages 300 RSS feeds, your twitter account, Facebook and everthing else then you’ll be fine. In fact your’re probably already there. For those of you who are looking for something a little less high intensity the following advice may be helpful.
- Pick a small amount of your existing feeds as a starting point to see what you feel comfortable with sharing. Be aware that if you share e.g. Flickr or YouTube feeds it will also include your favourites, including old ones. Do share something – even if only some links – otherwise people won’t know that you’re there.
- Subscribe to someone you know and trust and stick with just one or two people for a while as you get to understand how things work. As you see extra stuff coming in from other people (friends of your friends) start to subscribe to one or two of them that you think look interesting. Do not subscribe to Robert Scoble if you don’t want to get swamped.
- Use the hide button. You probably don’t need to know about everyone’s favourite heavy metal bands (or perhaps you do). The hide button can get rid of a specific service from a specific person but you can set it so that you do so it if other people like it.
- Don’t worry if you can’t keep up. Using the Best of the Day/Week/Month button will let you catch up on what people thought was important.
- Find a schedule that suits you and stick to it. While the current users are dominated by the ‘always on’ brigade that doesn’t mean you need to do it the same way. But also don’t feel that because you came in late you can’t comment. It may just be that you are needed to kick that conversation back onto some people’s front page
- Join the Life Scientists Room and share interesting stuff. This provides a place to put particularly interesting links and is followed by a fair number of people, probably more than you are. If it is worthy of comment then put it in front of people. If you aren’t sure whether its relevant ask, you can always start a new room if need be.
- Enjoy, comment and participate in a way you feel comfortable with. This is a (potential) work tool. If it works for you, great! If not well so be it – there’ll be another one along in a minute.
Data is free or hidden – there is no middle ground
Science commons and other are organising a workshop on Open Science issues as a satellite meeting of the European Science Open Forum meeting in July. This is pitched as an opportunity to discuss issues around policy, funding, and social issues with an impact on the ‘Open Research Agenda’. In preparation for that meeting I wanted to continue to explore some of the conflicts that arise between wanting to make data freely available as soon as possible and the need to protect the interests of the researchers that have generated data and (perhaps) have a right to the benefits of exploiting that data.
John Cumbers proposed the idea of a ‘Protocol’ for open science that included the idea of a ‘use embargo’; the idea that when data is initially made available, no-one else should work on it for a specified period of time. I proposed more generally that people could ask that people leave data alone for any particular period of time, but that there ought to be an absolute limit on this type of embargo to prevent data being tied up. These kinds of ideas revolve around the need to forge community norms – standards of behaviour that are expected, and to some extent enforced, by a community. The problem is that these need to evolve naturally, rather than be imposed by committee. If there isn’t community buy in then proposed standards have no teeth.
An alternative approach to solving the problem is to adopt some sort ‘license’. A legal or contractual framework that creates obligation about how data can be used and re-used. This could impose embargoes of the type that John suggested, perhaps as flexible clauses in the license. One could imagine an ‘Open data – six month analysis embargo’ license. This is attractive because it apparently gives you control over what is done with your data while also allowing you to make it freely available. This is why people who first come to the table with an interest in sharing content always start with CC-BY-NC. They want everyone to have their content, but not to make money out of it. It is only later that people realise what other effects this restriction can have.
I had rejected the licensing approach because I thought it could only work in a walled garden, something which goes against my view of what open data is about. More recently John Wilbanks has written some wonderfully clear posts on the nature of the public domain, and the place of data in it, that make clear that it can’t even work in a walled garden. Because data is in the public domain, no contractual arrangement can protect your ability to exploit that data, it can only give you a legal right to punish someone who does something you haven’t agreed to. This has important consequences for the idea of Open Science licences and standards.
If we argue as an ‘Open Science Movement’ that data is in and must remain in the public domain then, if we believe this is in the common good, we should also argue for the widest possible interpretation of what is data. The results of an experiment, regardless of how clever its design might be, are a ‘fact of nature’, and therefore in the public domain (although not necessarily publically available). Therefore if any person has access to that data they can do whatever the like with it as long as they are not bound by a contractual arrangement. If someone breaks a contractual arrangement and makes the data freely available there is no way you can get that data back. You can punish the person who made it available if they broke a contract with you. But you can’t recover the data. The only way you can protect the right to exploit data is by keeping it secret. The is entirely different to creative content where if someone ignores or breaks licence terms then you can legally recover the content from anyone that has obtained it.
Why does this matter to the Open Science movement? Aren’t we all about making the data available for people to do whatever anyway? It matters because you can’t place any legal limitations on what people do with data you make available. You can’t put something up and say ‘you can only use this for X’ or ‘you can only use it after six months’ or even ‘you must attribute this data’. Even in a walled garden, once there is one hole, the entire edifice is gone. The only way we can protect the rights of those who generate data to benefit from exploiting it is through the hard work of developing and enforcing community norms that provide clear guidelines on what can be done. It’s that or simply keep the data secret.
What is important is that we are clear about this distinction between legal and ethical protections. We must not tell people that their data can be protected because essentially they can’t. And this is a real challenge to the ethos of open data because it means that our only absolutely reliable method for protecting people is by hiding data. Strong community norms will, and do, help but there is a need to be careful about how we encourage people to put data out there. And we need to be very strong in condemning people who do the ‘wrong’ thing. Which is why a discussion on what we believe is ‘right’ and ‘wrong’ behaviour is incredibly important. I hope that discussion kicks off in Barcelona and continues globally over the next few months. I know that not everyone can make the various meetings that are going on – but between them and the blogosphere and the ‘streamosphere‘ we have the tools, the expertise, and hopefully the will, to figure these things out.
The trouble with institutional repositories
 I spent today at an interesting meeting at Talis headquarters where there was a wide range of talks. Most of the talks were liveblogged by Andy Powell and also by Owen Stephens (who has written a much more comprehensive summary of Andy’s talk) and there will no doubt be some slides and video available on the web in future. The programme is also available. Here I want to focus on Andy Powell’s talk (slides), partly because he obviously didn’t liveblog it but primarily because it crystallised for me many aspects of the way we think about Institutional Repositories. For those not in the know, these are warehouses that are becoming steadily more popular, run generally by unversities to house their research outputs, in most cases peer reviewed papers. Self archiving of some version of published papers is the so called ‘Green Route’ to open access.
I spent today at an interesting meeting at Talis headquarters where there was a wide range of talks. Most of the talks were liveblogged by Andy Powell and also by Owen Stephens (who has written a much more comprehensive summary of Andy’s talk) and there will no doubt be some slides and video available on the web in future. The programme is also available. Here I want to focus on Andy Powell’s talk (slides), partly because he obviously didn’t liveblog it but primarily because it crystallised for me many aspects of the way we think about Institutional Repositories. For those not in the know, these are warehouses that are becoming steadily more popular, run generally by unversities to house their research outputs, in most cases peer reviewed papers. Self archiving of some version of published papers is the so called ‘Green Route’ to open access.
The problem with institutional repositories in their current form is that academics don’t use them. Even when they are being compelled there is massive resistance from academics. There are a variety of reasons for this: academics don’t like being told how to do things; they particularly don’t like being told what to do by their institution; the user interfaces are usually painful to navigate. Nonetheless they are a valuable part of the route towards making more research results available. I use plenty of things with ropey interfaces because I see future potential in them. Yet I don’t use either of the repositories in the places where I work – in fact they make my blood boil when I am forced to. Why?
So Andy was talking about the way repositories work and the reasons why people don’t use them. He had already talked about the language problem. We always talk about ‘putting things in the repository’ rather than ‘making them available on the web’. He had mentioned already that the institutional nature of repositories does not map well onto the social networks of the academic users which probably bear little relationship with institutions and are much more closely aligned to discipline and possibly geographic boundaries (although they can easily be global).
But for me the key moment was when Andy asked ‘How many of you have used SlideShare’. Half the people in the room put their hands up. Most of the speakers during the day pointed to copies of their slides on SlideShare. My response was to mutter under my breath ‘And how many of them have put presentations in the institutional repository?’ The answer to this; probably none. SlideShare is a much better ‘repository’ for slide presentations than IRs. There are more there, people may find mine, it is (probably) Google indexed. But more importantly I can put slides up with one click, it already knows who I am, I don’t need to put in reams of metadata, just a few tags. And on top of this it provides added functionality including embedding in other web documents as well as all the social functions that are a natural part of a ‘Web2.0’ site.
SlideShare is a very good model of what a Repository can be. It has issues. It is a third party product, it may not have long term stability, it may not be as secure as some people would like. But it provides much more of the functionality that I want from a service for making my presentations available on the web. It does not serve the purpose of an archive – and maybe an institutional repository is better in that role. But for the author, the reason for making things available is so that people use them. If I make a video that relates to my research it will go on YouTube, Bioscreencast, or JoVE, not in the institutional repository, I put research related photos on Flickr, not in the institutional repository, and critically, I leave my research papers on the websites of the journal that published them, and cannot be bothered with the work required to put them in the institutional repository.
Andy was arguing for global discipline specific repositories. I would suggest that the lesson of the Web2.0 sites is that we should have data type specific repositories. FlickR is for pictures, SlideShare for presentations. In each case the specialisation enables a sort of implicit metadata and for the site to concentrate on providing functionality that adds value to that particular data type. Science repositories could win by doing the same. PDB, GenBank, SwissProt deal with specific types of data. Some might argue that GenBank is breaking under the strain of the different types and quantities of data generated by the new high throughput sequencing tools. Perhaps a new repository is required that is specially designed for this data.
So what is the role for the institutional repository? The preservation of data is one aspect. Pulling down copies of everything to provide an extra backup and retain an institutional record. If not copying then indexing and aggregating so as to provide a clear guide to the institutions outputs. This needn’t be handled in house of course and can be outsourced. As Paul Miller suggested over lunch, the role of the institution need not be to keep a record of everything, but to make sure that such a record is kept. Curation may be another, although that may be too big a job to be tackled at institutional level. When is a decision made that something isn’t worth keeping anymore? What level of metadata or detail is worth preserving?
But the key thing is that all of this should be done automatically and must not require intervention by the author. Nothing drives me up the wall more than having to put the same set of data into two subtly different systems more than once. And as far as I can see there is no need to do so. Aggregate my content automatically, wrap it up and put it in the repository, but I don’t want to have to deal with it. Even in the case of peer reviewed papers it ought to be feasible to pull down the vast majority of the metadata required. Indeed, even for toll access publishers, everything except the appropriate version of the paper. Send me a polite automated email and ask me to attach that and reply. Job done.
For this to really work we need to take an extra step in the tools available. We need to move beyond files that are simply ‘born digital’ because these files are in many ways still born. This current blog post, written in Word on the train is a good example. The laptop doesn’t really know who I am, it probably doesn’t know where I am, and it has not context for the particular word document I’m working on. When I plug this into the WordPress interface at OpenWetWare all of this changes. The system knows who I am (and could do that through OpenID). It knows what I am doing (writing a Blog post) and the Zemanta Firefox plug in does much better than that, suggesting tags, links, pictures and keywords.
Plugins and online authoring tools really have the potential to automatically generate those last pieces of metadata that aren’t already there. When the semantics comes baked in then the semantic web will fly and the metadata that everyone knows they want, but can’t be bothered putting in, will be available and re-useable, along with the content. When documents are not only born digital but born on and for the web then the repositories will have probably still need to trawl and aggregate. But they won’t have to worry me about it. And then I will be a happy depositor.
Defining error rates in the Illumina sequence: A useful and feasible open project?
Regular readers will know I am a great believer in the potential of Web2.0 tools to enable rapid aggregation of loose networks of collaborators to solve a particular problem and the possibilities of using this approach to do science better, faster, and more efficiently. The reason why we haven’t had great successes on this thus far is fundamentally down to the size of the network we have in place and the bias in the expertise of that network towards specific areas. There is a strong bioinformatics/IT bias in the people interested in these tools and this plays out in a number of fields from the people on Friendfeed, to the relative frequency of commenting on PLoS Computational Biology versus PLoS ONE.
Putting these two together one obvious solution is to find a problem that is well suited to the people who are around, may be of interest to them, and is also quite useful to solve. I think I may have found such a problem.
The Illumina next generation sequencing platform developed originally by Solexa is the latest kid on the block as far as the systems that have reached the market. I spent a good part of today talking about how the analysis pipeline for this system could be improved. But one thing that came out as an issue is that no-one seems to have published detailed analysis of the types of errors that are generated experimentally by this system. Illumina probably have done this analysis in some form but have better things to do than write it up.
The Solexa system is based on sequencing by synthesis. A population of DNA molecules, all amplified from the same single molecule, is immobilised on a surface. A new strand of DNA is added, one base at a time. In the Solexa system each base has a different fluorescent marker on it plus a blocking reagent. After the base is added, and the colour read, the blocker is removed and the next base can be added. More details can be found on the genographia wiki. There are two major sources of error here. Firstly, for a proportion of each sample, the base is not added successfully. This means in the next round, that part of the sample may generate a readout for the previous base. Secondly the blocker may fail, leading to the addition of two bases, causing a similar problem but in reverse. As the cycles proceed the ends of each DNA strand in the sample get increasingly out of phase making it harder and harder to tell which is the correct signal.
These error rates are probably dependent both on the identity of the base being added and the identity of the previous base. It may also be related to the number of cycles that have been carried out. There is also the possibility that the sample DNA has errors in it due to the amplification process though these are likely to be close to insignificant. However there is no data on these error rates available. Simple you might think to get some of the raw data and do the analysis – fit the sequence of raw intensity data to a model where the parameters are error rates for each base.
Well we know that the availability of data makes re-processing possible and we further believe in the power of the social network. And I know that a lot of you guys are good at this kind of analysis, and might be interested in having a play with some of the raw data. It could also be a good paper – Nature Biotech/Nature Methods perhaps and I am prepared to bet it would get an interesting editorial writeup on the process as well. I don’t really have the skills to do the work but if others out there are interested then I am happy to coordinate. This could all be done, in the wild, out in the open and I think that would be a brilliant demonstration of the possibilities.
Oh, the data? We’ve got access to the raw and corrected spot intensities and the base calls from a single ‘tile’ of the phiX174 control lane for a run from the 1000 Genomes Project which can be found at http://sgenomics.org/phix174.tar.gz courtesy of Nava Whiteford from the Sanger Centre. If you’re interested in the final product you can see some of the final read data being produced here.
What I had in mind was taking the called sequence, align onto phiX174 so we know the ‘true’ sequence. Then use that sequence plus a model with error rates to parameterise those error rates. Perhaps there is a better way to approach the problem? There are a series of relatively simple error models that could be tried and if the error rates can be defined then it will enable a really significant increase in both the quality and quantity of data that can be determined by these machines. I figure splitting the job up into a few small groups working on different models, putting the whole thing up on google code with a wiki there to coordinate and capture other issues as we go forward. Anybody up for it (and got the time)?
Twittering labs? That is just so last year…

The Mars Phoenix landing has got a lot of coverage around the web, particularly from some misty eyed old blokes who remember watching landings via the Mosaic browser in an earlier, simpler age. The landing is cool, but one thing I thought was particularly clever was the use of Twitter by JPL to publicise the landing and what is happening on a minute to minute basis. Now my suspicion is that they haven’t actually installed Twhirl on the Phoenix Lander and that there is actually a person at JPL writing the Tweets. But that isn’t the point. The point is that the idea of an instrument (or in this case a spacecraft) outputting a stream of data is completely natural to people. The idea of the overexcited lander saying ‘come on rocketsssssss!!!!!!!!’ is very appealing (you can tell it’s a young spaceship, it hasn’t learnt not to shout yet; although if your backside was at 2,000 °C you might have something to say about it as well).
I’ve pointed out some cool examples of this in the past including London Bridge and Steve Wilson, in Jeremy Frey’s group at Southampton, has been doing some very fun stuff both logging what happens in a laboratory, and blogging that out to the web, using the tools developed by the Simile team at MIT. The notion of the instrument generating a data stream and using that stream as an input to an authoring tool like a laboratory notebook or into other automated processes is a natural one that fits well both with the way we work in the laboratory (even when your laboratory is the solar system) and our tendency to anthropomorphise our kit. However, the day the FPLC tells me it had a hard night and doesn’t feel like working this morning is the day it gets tossed out. And the fact that it was me that fed it the 20% ethanol is neither here nor there.
Now the question is; can I persuade JPL to include actual telemetry, command, and acknowledgement data in the twitter stream? That would be very cool.
How do we build the science data commons? A proposal for a SciFoo session
 I realised the other day that I haven’t written an exciteable blog post about getting an invitation to SciFoo! The reason for this is that I got overexcited over on FriendFeed instead and haven’t really had time to get my head together to write something here. But in this post I want to propose a session and think through what the focus and aspects of that might be.
I realised the other day that I haven’t written an exciteable blog post about getting an invitation to SciFoo! The reason for this is that I got overexcited over on FriendFeed instead and haven’t really had time to get my head together to write something here. But in this post I want to propose a session and think through what the focus and aspects of that might be.
I am a passionate advocate of two things that I think are intimately related. I believe strongly in the need and benefits that will arise from building, using, and enabling the effective search and processing of a scientific data commons. I [1,2] and others (including John Wilbanks, Deepak Singh, and Plausible Accuracy) have written on this quite a lot recently. The second aspect is that I believe strongly in the need for effective useable and generic tools to record science as it happens and to process that record so that others can use it effectively. To me these two things are intimately related. By providing the tools that enable the record to be created and integrating them with the systems that will store and process the data commons we can enable scientists to record their work better, communicate it better, and make it available as a matter of course to other scientists (not necessarily immediately I should add, but when they are comfortable with it). Continue reading “How do we build the science data commons? A proposal for a SciFoo session”
Approaching deadline for Open Science@PSB
Just a gentle reminder that the deadline for submissions for the Open Science Workshop at Pacific Symposium on Biocomputing is approaching. The purpose of the early deadline is so that we can give people plenty of notice that they have a talk so they or we can sort out funding. At this stage we really only need an abstract or even an outline so we can organise the program. We are hoping to be able to make a contribution to the costs of speakers and poster presenters so if you will need or would appreciate support then please make a note of that in your submission. And please do get in touch if you want to come but money is an issue and we will work with you to see what we can. If there is enough UK/Europe interest we can look at putting in a small grant application. The US seems a bit harder but we are working on that as well. As Shirley Wu has mentioned we are actively pursuing a range of fundrasing options (watch this space for more t-shirts and other open science merchandise – and yes we are doing more than just designing t-shirts). Any help with contacts or cold hard cash will also be greatly appreciated.
It is looking like it will be an exciting programme and we would like as many people to be there as possible. Submission instructions are at the Call for Proposals. You know you want to spend the middle of January in Hawaii, so this is the excuse you’ve been looking for.