Tomorrow I am giving a talk at the UKOLN Institutional Web Managers workshop starting at 12:45 British Summer Time (GMT+1). In principle you will be able to see the talk video cast at the links on the video streaming page. The page also has a liveblogging tool (OpenID enabled apparently!). I won’t be liveblogging my own talk but I will be attempting to respond to comments or questions either on that tool, via FriendFeed, or @cameronneylon on Twitter. I make no promises that this will work but if it all fails then I will record a live screencast.
Policy for Open Science – reflections on the workshop
Written on the train on the way from Barcelona to Grenoble. This life really is a lot less exotic than it sounds…Â
The workshop that I’ve reported on over the past few days was both positive and inspiring. There is a real sense that the ideas of Open Access and Open Data are becoming mainstream. As several speakers commented, within 12-18 months it will be very unusual for any leading institution not to have a policy on Open Access to its published literature. In many ways as far as Open Access to the published literature is concerned the war has been won. There will remains battles to be fought over green and gold routes – the role of licenses and the need to be able to text mine – successful business models remain to be made demonstrably sustainable – and there will be pain as the inevitable restructuring of the publishing industry continues. But be under no illusions that this restructuring has already begun and it will continue in the direction of more openness as long as the poster children of the movement like PLoS and BMC continue to be successful.
Open Data remains further behind, both with respect to policy and awareness. Many people spoke over the two days about Open Access and then added, almost as an addendum ‘Oh and we need to think about data as well’. I believe the policies will start to grow and examples such as the BBSRC Data Sharing Policy give a view of the future. But there is still much advocacy work to be done here. John Wilbanks talked about the need to set achievable goals, lines in the sand which no-one can argue with. And the easiest of these is one that we have discussed many times. All data associated with a published paper, all analysis, and all processing procedures, should be made available. This is very difficult to argue with – nonetheless we know of examples where the raw data of important experiments is being thrown away. But if an experiment cannot be shown to have been done, cannot be replicated and checked, can it really be publishable? Nonetheless this is a very useful marker, and a meme that we can spread and promote.
In the final session there was a more critical analysis of the situation. A number of serious questions were raised but I think they divide into two categories. The first involves the rise of the ‘Digital Natives’ or the ‘Google Generation’. The characteristics of this new generation (a gross simplification in its own right) are often presented as a pure good. Better networked, more sharing, better equipped to think in the digital network. But there are some characteristics that ought to give pause. A casualness about attribution, a sense that if something is available then it is fine to just take it (its not stealing after all, just copying). There is perhaps a need to recover the roots of ‘Mertonian’ science, to as I think James Boyle put it, publicise and embed the attitudes of the last generation of scientists, for whom science was a public good and a discipline bounded by strict rules of behaviour. Some might see this as harking back to an elitist past but if we are constructing a narrative about what we want science to be then we can take the best parts of all of our history and use it to define and refine our vision. There is certainly a place for a return to the compulsory study of science history and philosophy.
The second major category of issues discussed in the last session revolved around the question of what do we actually do now. There is a need to move on many fronts, to gather evidence of success, to investigate how different open practices work – and to ask ourselves the hard questions. Which ones do work, and indeed which ones do not. Much of the meeting revolved around policy with many people in favour of, or at least not against, mandates of one sort or another. Mike Carroll objected to the term mandate – talking instead about contractual conditions. I would go further and say that until these mandates are demonstrated to be working in practice they are aspirations. When they are working in practice they will be norms, embedded in the practice of good science. The carrot may be more powerful than the stick but peer pressure is vastly more powerful than both.
So they key questions to me revolve around how we can convert aspirations into community norms. What is needed in terms of infrastructure, in terms of incentives, and in terms of funding to make this stuff happen? One thing is to focus on the infrastructure and take a very serious and critical look at what is required. It can be argued that much of the storage infrastructure is in place. I have written on my concerns about institutional repositories but the bottom line remains that we probably have a reasonable amount of disk space available. The network infrastructure is pretty good so these are two things we don’t need to worry about. What we do need to worry about, and what wasn’t really discussed very much in the meeting, is the tools that will make it easy and natural to deposit data and papers.
The incentive structure remains broken – this is not a new thing – but if sufficiently high profile people start to say this should change, and act on those beliefs, and they are, then things will start to shift. It will be slow but bit by bit we can imagine getting there. Can we take shortcuts. Well there are some options. I’ve raised in the past the idea of a prize for Open Science (or in fact two, one for an early career researcher and one for an established one). Imagine if we could make this a million dollar prize, or at least enough for someone to take a year off. High profile, significant money, and visible success for someone each year. Even without money this is still something that will help people – give them something to point to as recognition of their contribution. But money would get people’s attention.
I am sceptical about the value of ‘microcredit’ systems where a person’s diverse and perhaps diffuse contributions are aggregated together to come up with some sort of ‘contribution’ value, a number by which job candidates can be compared. Philosophically I think it’s a great idea, but in practice I can see this turning into multiple different calculations, each of which can be gamed. We already have citation counts, H-factors, publication number, integrated impact factor as ways of measuring and comparing one type of output. What will happen when there are ten or 50 different types of output being aggregated? Especially as no-one will agree on how to weight them. What I do believe is that those of us who mentor staff, or who make hiring decisions should encourage people to describe these contributions, to include them in their CVs. If we value them, then they will value them. We don’t need to compare the number of my blog posts to someone else’s – but we can ask which is the most influential – we can compare, if subjectively, the importance of a set of papers to a set of blog posts. But the bottom line is that we should actively value these contributions – let’s start asking the questions ‘Why don’t you write online? Why don’t you make your data available? Where are your protocols described? Where is your software, your workflows?’
Funding is key, and for me one of the main messages to come from the meeting was the need to think in terms of infrastructure, and in particular, to distinguish what is infrastructure and what is science or project driven. In one discussion over coffee I discussed the problem of how to fund development projects where the two are deeply intertwined and how this raises challenges for funders. We need new funding models to make this work. It was suggested in the final panel that as these tools become embedded in projects there will be less need to worry about them in infrastructure funding lines. I disagree. Coming from an infrastructure support organisation I think there is a desperate need for critical strategic oversight of the infrastructure that will support all science – both physical facilities, network and storage infrastructure, tools, and data. This could be done effectively using a federated model and need not be centralised but I think there is a need to support the assumption that the infrastructure is available and this should not be done on a project by project basis. We build central facilities for a reason – maybe the support and development of software tools doesn’t fit this model but I think it is worth considering.
This ‘infrastructure thinking’ goes wider than disk space and networks, wider than tools, and wider than the data itself. The concept of ‘law as infrastructure’ was briefly discussed. There was also a presentation looking at different legal models of a ‘commons’; the public domain, a contractually reconstructed commons, escrow systems etc. In retrospect I think there should have been more of this. We need to look critically at different models, what they are good for, how they work. ‘Open everything’ is a wonderful philosophical position but we need to be critical about where it will work, where it won’t, and where it needs contractual protection, or where such contractual protection is counter productive. I spoke to John Wilbanks about our ideas on taking Open Source Drug Discovery into undergraduate classes and schools and he was critical of the model I was proposing, not from the standpoint of the aims or where we want to be, but because it wouldn’t be effective at drawing in pharmaceutical companies and protecting their investment. His point was, I think, that by closing off the right piece of the picture with contractual arrangements you bring in vastly more resources and give yourself greater ability to ensure positive outcomes. That sometimes to break the system you need to start by working within it by, in this case, making it possible to patent a drug. This may not be philosophically in tune with my thinking but it is pragmatic. There will be moments, especially when we deal with the interface with commerce, where we have to make these types of decisions. There may or may not be ‘right’ answers, and if there are they will change over time but we need to know our options and know them well so as to make informed decisions on specific issues.
But finally, as is my usual wont, I come back to the infrastructure of tools. The software that will actually allow us to record and order this data that we are supposed to be sharing. Again there was relatively little on this in the meeting itself. Several speakers recognised the need to embed the collection of data and metadata within existing workflows but there was very little discussion of good examples of this. As we have discussed before this is much easier for big science than for ‘long tail’ or ‘small science’. I stand by my somewhat provocative contention that for the well described central experiments of big science this is essentially a solved problem – it just requires the will and resources to build the language to describe the data sets, their formats, and their inputs. But the problem is that even for big science, the majority of the workflow is not easily automated. There are humans involved, making decisions moment by moment, and these need to be captured. The debate over institutional repositories and self archiving of papers is instructive here. Most academics don’t deposit because they can’t be bothered. The idea of a negative click repository – where this is a natural part of the workflow can circumvent this. And if well built it can make the conventional process of article submission easier. It is all a question of getting into the natural workflow of the scientist early enough that not only do you capture all the contextual information you want, but that you can offer assistance that makes them want to put that information in.
The same is true for capturing data. We must capture it at source. This is the point where it has the potential to add the greatest value to the scientist’s workflow by making their data and records more available, by making them more consistent, by allowing them to reformat and reanalyse data with ease, and ultimately by making it easy for them to share the full record. We can and we will argue about where best to order and describe the elements of this record. I believe that this point comes slightly later – after the experiment – but wherever it happens it will be made much easier by automatic capture systems that hold as much contextual information as possible. Metadata is context – almost all of it should be possible to catch automatically. Regardless of this we need to develop a diverse ecosystem of tools. It needs to be an open and standards based ecosystem and in my view needs to be built up of small parts, loosely coupled. We can build this – it will be tough, and it will be expensive but I think we know enough now to at least outline how it might work, and this is the agenda that I want to explore at SciFoo.
John Wilbanks had the last word, and it was a call to arms. He said ‘We are the architects of Open’. There are two messages in this. The first is we need to get on and build this thing called Open Science. The moment to grasp and guide the process is now. The second is that if you want to have a part in this process the time to join the debate is now. One thing that was very clear to me was that the attendees of the meeting were largely disconnected from the more technical community that reads this and related blogs. We need to get the communication flowing in both directions – there are things the blogosphere knows, that we are far ahead on, and we need to get the information across. There are things we don’t know much about, like the legal frameworks, the high level policy discussions that are going on. We need to understand that context. It strikes me though that if we can combine the strengths of all of these communities and their differing modes of communication then we will be a powerful force for taking forward the open agenda.
Policy for open science – the wrap up session
Today Science Commons sponsored a meeting looking at the policy issues involved in Open Access and Open Science more widely. I blogged James Boyle’s keynote earlier and there was some notes along the way via Twitter. This is a set of notes from the last session of the meeting, a panel with Alexis-Michel Mugabushaka (European Science Foundation), Javier Hernandez-Ros (European Commission), Michael Carroll (University of Villanova, Creative Commons) and John Wilbanks (Science Commons). These notes were taken at speed and are my own record of what happened. They should not necessarily be taken as a transcript of what the panellists said and any inaccuracies are my own fault.
It is important to focus on the barriers to open science. Much has been said about ideals and beliefs but there remains a paucity of real evidence to support the assertions of the open science community. Changing policy, culture, and challenging entrenched positions, putting 50,000 jobs in the publishing industry at risk, requires strong evidence of benefits. On top of this the consideration of efficiency both with respect to time and money. Details are important. What is data? What is meant by data sharing? Both from a legal and descriptive perspective.
Some issues looked at by the European Commission:
1) Who does what, how is this to be organised? At national, institutional, discpline, European, level? A mixture of top down and bottom up witll be required to make progress. Again efficient provision is required because uneceessary duplication wll be counterproductive and expensive.
2)Â The legal issues, copright etc. The issue is not copyright per se but licensing and the power games associated with the ecomonic need of commercial entities. There is a need for a meaningful discussion across the stakeholders.Â
3) Technical issues. The whole area is developing and it is not clear where the funding streams will continue as these become more embedded in general scientific practice.
The key issues are to bring everyone on board as part of the discussion. What is the contribution that publishers can make? View this from a positive angle, not a a combative angle. What are the roles of the various stakeholders.
Things actually need to be done! Much talk about what should be done. Less on how to actually do it.
This meeting is a momentin time where the idea of Openness has become mainstream. The argument started on the fringe. There is a risk of over confidence in the arguments. The issue of evidence is an important one. Can ROI be actually shown? It is known that there are unexpected audiences for information from the web – new innovation will emerge but it is unclear how much or where it will come from.
In the short term there is still a fight but in the long term the world will change – as the next generation of scientists come through. But we don’t necessarily need or want to wait for them to arrive. We may want to ensure that some values from the previous generation of scientists are transmitted effectively. This moment in time is still transitional. Most of us were born into an analogue world but we still don’t think digitally. When people design initiatives and projects with the network in mind – rather than as an afterthought or add on – the efficiencies will be seen much more strongly. There is an argument that the EU model of funding and thinking can help to drive the network effects that will demonstrate the benefits more effectively than the US funding models. A missing part is the link to business models – what is the profit model for openness? Again, Google works (and is profitable).
The fight will go on – but thinking forward, how would we do work and plan for future projects built into the network.
The law as infrastructure. Creative Commons came out of the wish of lawyers to build a technical infrastructure that works for open networks. The legal issues of sharing data haven’t been discussed very much. How can a legal infrastructure be built to assist openness. This is the role of Creative/Science Commons. ‘No-one want to invite lawyers to the party’ but some of them really want to help it go off well!
The purpose of the meeting was to bring people together. There is a need for people to understand they are doing the same thing in different domains. It is difficult to fund the commons – but by brnging people together it is possible to build a social network that may assist in identifying funding options. Science funders don’t do policy, but policy people don’t fund science. A policy statement will be forthcoming in the future taking the current recommendations.
The benefits of the open web come from the explosion of people actually using a computer network. We must think of the users of an open architected science with the same potential for explosion. Can we make it possible to do science withouot being a) rich and b) a member of a closed guild. What would happen if 100 million people could ask scientific questions – even if relatively few of them were really smart questions.
The tower of babel is all over science but also all over computation. What we need is an institute of science langugages to standardise. A cancer marker with 94 synonyms would make Academic Francaise apoplectic. We are a long way from being able to solve either of these problems. How can the tools and standards be built.
We are the architects of Open. We need to link all the work that has been described in the meeting. We need an Open architecture to describe and deliver openness. The short term impact of technology is always over estimated but the long term is underestimated. We don’t have to worry too much about the long term impacr – we can just get on and do it.
‘In theory there is no difference between theory and practice. In practice there is’ – lets just deploy and get on with it. Lets build the evidence and the tools and the standards that will take us forward.
Policy and technology for e-science – A forum on on open science policy
I’m in Barcelona at a satellite meeting of the EuroScience Open Forum organised by Science Commons and a number of their partners. Today is when most of the meeting will be with forums on ‘Open Access Today’, ‘Moving OA to the Scientific Enterprise:Data, materials, software’, ‘Open access in the the knowledge network’, and ‘Open society, open science: Principle and lessons from OA’. There is also a keynote from Carlos Morais-Pires of the European Commission and the lineup for the panels is very impressive.
Last night was an introduction and social kickoff as well. James Boyle (Duke Law School, Chair of board of directors of Creative Commons, Founder of Science commons) gave a wonderful talk (40 minutes, no slides, barely taking breath) where his central theme was the relationship between where we are today with open science and where international computer networks were in 1992. He likened making the case for open science today with that of people suggesting in 1992 that the networks would benefit from being made freely accessible, freely useable, and based on open standards. The fears that people have today of good information being lost in a deluge of dross, of their being large quantities of nonsense, and nonsense from people with an agenda, can to a certain extent be balanced against the idea that to put it crudely, that Google works. As James put it (not quite a direct quote) ‘You need to reconcile two statements; both true. 1) 99% of all material on the web is incorrect, badly written, and partial. 2) You probably haven’t opened an encylopedia as a reference in ten year.
James gave two further examples, one being the availability of legal data in the US. Despite the fact that none of this is copyrightable in the US there are thriving businesses based on it. The second, which I found compelling, for reasons that Peter Murray-Rust has described in some detail. Weather data in the US is free. In a recent attempt to get long term weather data a research effort was charged on the order of $1500, the cost of the DVDs that would be needed to ship the data, for all existing US weather data. By comparison a single German state wanted millions for theirs. The consequence of this was that the European data didn’t go into the modelling. James made the point that while the European return on investment for weather data was a respectable nine-fold, that for the US (where they are giving it away remember) was 32 times. To me though the really compelling part of this argument is if that data is not made available we run the risk of being underwater in twenty years with nothing to eat. This particular case is not about money, it is potentially about survival.
Finally – and this you will not be surprised was the bit I most liked – he went on to issue a call to arms to get on and start building this thing that we might call the data commons. The time has come to actually sit down and start to take these things forward, to start solving the issues of reward structures, of identifying business models, and to build the tools and standards to make this happen. That, he said was the job for today. I am looking forward to it.
I will attempt to do some updates via twitter/friendfeed (cameronneylon on both) but I don’t know how well that will work. I don’t have a roaming data tariff and the charges in Europe are a killer so it may be a bit sparse.
Science in the YouTube Age – introductory screencast for a talk I’m giving at IWMW
The UKOLN Institutional Web Managers Workshop is running in Aberdeen from 22-24 July and I am giving a talk discussing the impact of Web2.0 tools on science. My main theme will be the that the main cultural reasons for lack of uptake relate to the fear of losing control over data and ideas. Web2.0 tools rely absolutely on the willingness of people to make useful material available. In science this material is data, ideas, protocols, and analyses. Prior to publication most scientists are very sceptical of making their hard earned data available – but without this the benefits that we know can be achieved through network effects, re-use of data, and critical analysis of data and analysis, will not be seen. The key to benefiting from web based technologies is adopting more open practices.
The video below is a screencast of a shorter version of the talk intended to give people the opportunity to make comments, ask questions, or offer suggestions. I wanted to keep it short so there are relatively few examples in there – there will be much more in the full talk. For those who can’t make it to Aberdeen I am told that the talks are expected to be live videocast and I will provide a URL as soon as I can. If this works I am also intending to try and respond to comments and questions via FriendFeed or Twitter in real time. This may be foolhardy but we’ll see how it goes. Web2 is supposed to be about real time interaction after all!
I don’t seem to be able to embed the video but you can find it here.
What I missed on my holiday or Why I like refereeing for PLoS ONE
I was away last week having a holiday and managed to miss the whole Declan Butler/PLoS/Blogosphere dustup. Looked like fun. I don’t want to add to the noise as I think there was a lot of knee jerk reactions and significantly more heat than light. For anyone coming here without having heard about this I will point at the original article, Bora’s summary of reactions, and Timo Hannay’s reply at Nature. What I wanted to add to the discussion was a point that I haven’t seen in my quick skimming of the whole debate (which is certainly not complete so if I missed this then please drop in a comment).
No-one as far as I can see has really twigged as to just how disruptive PLoS ONE really is. In this I agree with Timo, in that I think publishers, from BMC, to Elsevier, ACS and Nature Publishing Group itself, should be very worried about the impact that it will have and think very hard about what it means for their future business models. Where we disagree, I think, is that I find this very exciting and think that it shows the way towards a scientific publishing industry that will look very different from todays’. Diffentiating on quality prior to publication was always difficult, and certainly expensive. The question for the future is whether we are prepared to pay for it, and are we getting value for money?
The criticism levelled at PLoS ONE is that it uses a ‘light touch’ refereeing process with the only criterion for publication being that a paper is methodologically sound. This, it is implied leads to a ‘low quality journal’ or perhaps rather a journal with a large number of relatively uncited articles. However there are very strong positives to this ‘light touch’ approach. It is fast. And it is cheap. The issue here is business models and the business model of PLoS ONE is highly disruptive. And financially successful. To me this is the big news. People are flocking to PLoS ONE because it is a quick and straightforward way of getting interesting (but perhaps not career making) results out there.
From an author’s perspective PLoS ONE cuts out the crap in getting papers published. The traditional approach (send to Nature/Science/Cell, get rejected, send to Nature/Science/Cell baby journal, get rejected, send to top tier specific journal, get rejected, end up eventually going to a journal that no-one subscribes to) takes time and effort and by the time you win someone else has usually published it anyway. It also costs the authors money in staff time to re-format, rejig, appease referees, re-jig again to appease a different set of referees. I haven’t done the sums but worst case scenario this could probably cost as much as a PLoS ONE publication charge. Save time, save money, still get indexed in PubMed. It starts to sound good, especially for all that material that you are not quite sure where to pitch.
But what about that stuff that is really hard hitting? That you know is important. Here you now have an interesting choice. You can send to Nature/Science/Cell/PLoS Biology and if you get past the initial editorial review stage and get to referees then you are probably looking at around six to nine months before publication. You will be in a high profile journal, can generate good publicity, have great paper on your CV. Alternatively you can send to PLoS ONE and have it on the web and in PubMed in perhaps two to four weeks. If the paper is as strong as you believe then you will still get your hundreds of citations, still have a great paper, still get good publicity. It probably doesn’t look quite so good on the traditional CV, but try putting the number of citations for each paper on your CV – that puts it in perspective. And it will be out a lot faster, you will be ahead of the game and you can apply for your next grant with ‘paper published and already cited three times’ not ‘paper submitted’ (read ‘about to be rejected’, there is an art in submitting papers just before the grant deadline). This makes for an interesting choice and one which cuts directly across the usual high impact/low impact criterion. It puts speed and convenience on the table as market differentiators in a way they haven’t been before.
As a referee PLoS ONE has a lot of appeal as well. You are being asked a very specific question. I recently refereed one paper for PLoS ONE at the same time as one for another (fairly low impact) journal. The PLoS ONE paper was a very simple case, the methodological detail was exemplary; easy to read, clear, and detailed. You get the impression the authors took care over it, possibly because they knew that was what it would be judged on (it is of course entirely possible that this group just writes good papers). The other paper was a distinct case of salami slicing – but I was left with trying to figure out whether it had been cut too thin for this specific journal. This is not just a difficult judgement to make. It is a highly subjective and probably meaningless one. The data was still useful and publishable, just probably not in that specific journal. Which one do you think took me longer? And which one left me with a warm feeling?
What about the reader? There is a lot of interesting stuff in PLoS ONE. There is also a lot of dross. But why should that matter? I don’t look at the dross; I often don’t even know that it exists. I can’t remember the last time I actually looked at a a journal table of contents. It doesn’t matter to me whether a paper is in Nature, Science, PLoS ONE, or Journal of the society for some highly specific thing in some rather small place. If it is searchable, and I have access to it then that’s all I need. If it is not both of these then for me it simply does not exist. And I don’t judge the value or reliability of an article based on where it is, I judge the article on what it contains. PLoS ONE actually wins here because its hard focus on being ‘methodologically sound’ tends to lead referees and editors (as well as authors) to focus on this aspect.
To me the truly radical thing about PLoS ONE is that is has redefined the nature of peer review and that people have bought into this model. The idea of dropping any assessment of ‘importance’ as a criterion for publication had very serious and very real risks for PLoS. It was entirely possible that the costs wouldn’t be usefully reduced. It was more than possible that authors simply wouldn’t submit to such a journal. PLoS ONE has successfully used a difference in its peer review process as the core of its appeal to its customers. The top tier journals have effectively done this for years at one end of the market. The success of PLoS ONE shows that it can be done in other market segments. What is more it suggests it can be done across existing market segments. That radical shift in the way scientific publishing works that we keep talking about? It’s starting to happen.
Live blogging at Friendfeed – Data sharing in the Biosciences: a sociological perspective
Open Science Workshop at Southampton – 31 August and 1 September 2008
Image via Wikipedia
I’m aware I’ve been trailing this idea around for sometime now but its been difficult to pin down due to issues with room bookings. However I’m just going to go ahead and if we end up meeting in a local bar then so be it! If Southampton becomes too difficult I might organise to have it at RAL instead but Southampton is more convenient in many ways.
Science Blogging 2008: London will be held on August 30 at the Royal Institution and as a number of people are coming to that it seemed a good opportunity to get a few more people together to have a get together and discuss how we might move things forward. This now turns out to be one of a series of such workshops following on from Collaborating for the future of open science, organised by Science Commons as a satellite meeting of EuroScience Open Forum in Barcelona next month, BioBarCamp/Scifoo from 5-10 August and a possible Open Science Workshop at Stanford on Monday 11 August, as well as the Open Science Workshop in Hawaii (can’t let the bioinformaticians have all the good conference sites to themselves!) at the Pacific Symposium on Biocomputing.
For the Southampton meeting I would propose that we essentially look at having four themed sessions: Tools, Data standards, Policy/Funding, and Projects. Within this we adopt an unconference style where we decide who speaks based on who is there and want to present something. My ideas is essentially to meet on the Sunday evening at a local hostelry to discuss and organise the specifics of the program for Monday. On the Monday we spend the day with presentations and leave plenty of room for discussion. People can leave in the afternoon, or hang around into the evening for further discussion. We have absolutely zero, zilch, nada funding available so I will be asking for a contribution (to be finalised later but probably £10-15 each) to cover coffee/tea and lunch on the Monday.
Travel plans
Image via Wikipedia
For anyone who is interested I thought it might be helpful to say where I am going to be at meetings and conferences over the next few months. If anyone is going then drop me a line and we can meet up. Most of these are looking like interesting meetings so I recommend going to them if they are in your area (and in your area if you see what I mean) Continue reading “Travel plans”
Friendfeed for scientists: What, why, and how?
There has been lots of interest amongst some parts of the community about what has been happening on FriendFeed. A growing number of people are signed up and lots of interesting conversations are happening. However it was suggested that as these groups grow they become harder to manage and the perceived barriers to entry get higher. So this is an attempt to provide a brief intro to FriendFeed for the scientist who may be interested in using it; what it is, why it is useful, and some suggestions on how to get involved without getting overwhelmed. This are entirely my views and your mileage may obviously vary.
What is FriendFeed?
FriendFeed is a ‘lifestreaming’ service or more simply a personal aggregator. It takes data streams that you generate and brings them all together into one place where people can see them. You choose to subscribe to any of the feeds you already generate (Flickr stream, blog posts, favorited YouTube videos, and lots of other services integrated). In addition you can post links to specific web pages or just comments into your stream. A number of these types of services have popped up in the recent months including Profilactic and Social Thing but FriendFeed has two key aspects that have led it to the fore. Firstly the commenting facilities enable rapid and effective conversations and secondly there was rapid adoption by a group of life scientists which has created a community. Like anything some of the other services have advantages and probably have their own communities but for science and in particular the life sciences FriendFeed is where it is at.
As well as allowing other people to look at what you have been doing FriendFeed allows you to subscribe to other people and see what they have been doing. You have the option of ‘liking’ particular items and commenting on them. In addition to seeing the items of your friends, people you are subscribed to, you also see items that they have liked or commented on. This helps you to find new people you may be interested in following. It also helps people to find you. As well as this items with comments or likes then get popped up to the top of the feed so items that are generating a conversation keep coming back to your attention.
These conversations can happen very fast. Some conversations baloon within minutes, most take place at a more sedate pace over a couple of hours or days but it is important to be aware that many people are live most of the time.
Why is FriendFeed useful?
So how is FriendFeed useful to a scientist? First and foremost it is a great way of getting rapid notification of interesting content from people you trust. Obviously this depends on there people who are interested in the same kinds of things that you are but this is something that will grow as the community grows. A number of FriendFeed users stream both del.icio.us bookmark pages as well as papers or web articles they have put into citeulike or connotea or simply via sharing it in Google Reader. Also you can get information that people have shared on opportunities, meetings, or just interesting material on the web. Think of it as an informal but continually running journal club – always searching for the next thing you will need to know about.
But FriendFeed is about much more than finding things on the web. One of its most powerful features is the conversations that can take place. Queries can be answered very rapidly going some way towards making possible the rapid formation of collaborative networks that can come together to solve a specific problem. Its not there yet but there are a growing number of examples where specific ideas were encouraged, developed, or problems solved quickly by bringing the right expertise to bear.
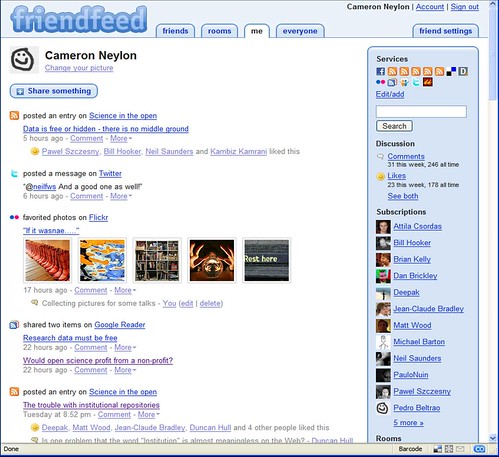
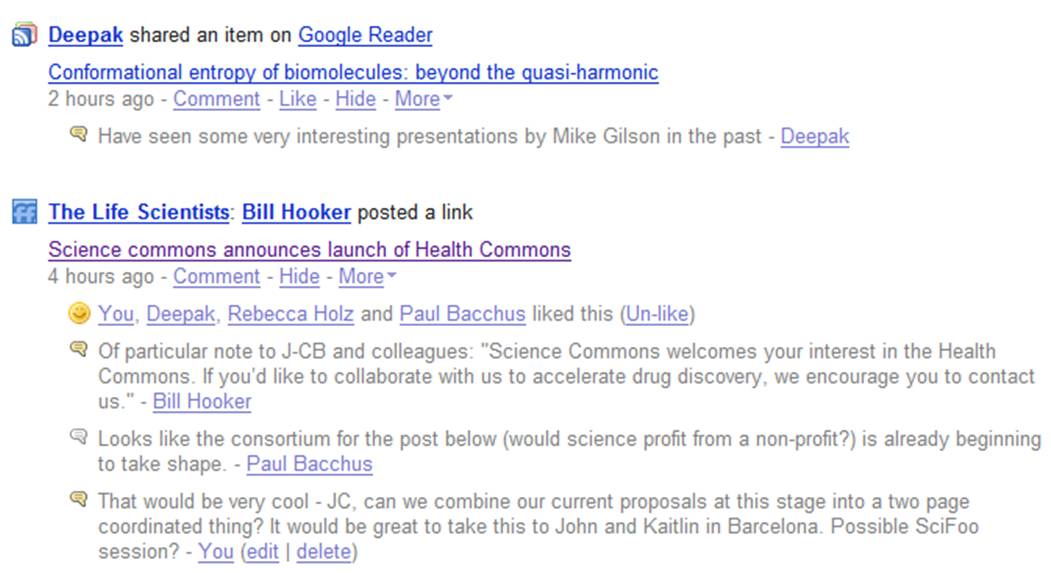
One example is shown in the following figure where I was looking for some help in building a particular protein model for a proposal. I didn’t really know how to go about this and didn’t have the appropriate software to hand. Pawel Szczesny offered to help and was able to quickly come up with what I wanted. In the future we hope to generate data which Pawel may be able to help us analyse. You can see the whole story and how it unfolded after this at http://friendfeed.com/search?q=mthk
We are still a long way from the dream of just putting out a request and getting an answer but it is worth point out that the whole exchange here lasted about four hours. Other collaborative efforts have also formed, most recently leading to the formation of BioGang, a collaborative area for people to work up and comment on possible projects.
So how do I use it? Will I be able to cope?
FriendFeed can be as high volume as you want it be but if its going to be useful to you it has to be manageable. If you’re the kind of person who already manages 300 RSS feeds, your twitter account, Facebook and everthing else then you’ll be fine. In fact your’re probably already there. For those of you who are looking for something a little less high intensity the following advice may be helpful.
- Pick a small amount of your existing feeds as a starting point to see what you feel comfortable with sharing. Be aware that if you share e.g. Flickr or YouTube feeds it will also include your favourites, including old ones. Do share something – even if only some links – otherwise people won’t know that you’re there.
- Subscribe to someone you know and trust and stick with just one or two people for a while as you get to understand how things work. As you see extra stuff coming in from other people (friends of your friends) start to subscribe to one or two of them that you think look interesting. Do not subscribe to Robert Scoble if you don’t want to get swamped.
- Use the hide button. You probably don’t need to know about everyone’s favourite heavy metal bands (or perhaps you do). The hide button can get rid of a specific service from a specific person but you can set it so that you do so it if other people like it.
- Don’t worry if you can’t keep up. Using the Best of the Day/Week/Month button will let you catch up on what people thought was important.
- Find a schedule that suits you and stick to it. While the current users are dominated by the ‘always on’ brigade that doesn’t mean you need to do it the same way. But also don’t feel that because you came in late you can’t comment. It may just be that you are needed to kick that conversation back onto some people’s front page
- Join the Life Scientists Room and share interesting stuff. This provides a place to put particularly interesting links and is followed by a fair number of people, probably more than you are. If it is worthy of comment then put it in front of people. If you aren’t sure whether its relevant ask, you can always start a new room if need be.
- Enjoy, comment and participate in a way you feel comfortable with. This is a (potential) work tool. If it works for you, great! If not well so be it – there’ll be another one along in a minute.