I’ve been interested for some time in capturing information and the context in which that information is created in the lab. The question of how to build an efficient and useable laboratory recording system is fundamentally one of how much information is necessary to record and how much of that can be recorded while bothering the researcher themselves as little as possible.
The Beyond the PDF mailing list has, since the meeting a few weeks ago, been partly focused on attempts to analyse human written text and to annotate these as structured assertions, or nanopublications. This is also the approach that many Electronic Lab Notebook systems attempt to take, capturing an electronic version of the paper notebook and in some cases trying to capture all the information in it in a structured form. I can’t help but feel that, while this is important, it’s almost precisely backwards. By definition any summary of a written text will throw away information, the only question is how much. Rather than trying to capture arbitrary and complex assertions in written text, it seems better to me to ask what simple vocabulary can be provided that can express enough of what people want to say to be useful.
In classic 80/20 style we ask what is useful enough to interest researchers, how much would we lose, and what would that be? This neatly sidesteps the questions of truth (though not of likelihood) and context that are the real challenge of structuring human authored text via annotation because the limited vocabulary and the collection of structured statements made provides an explicit context.
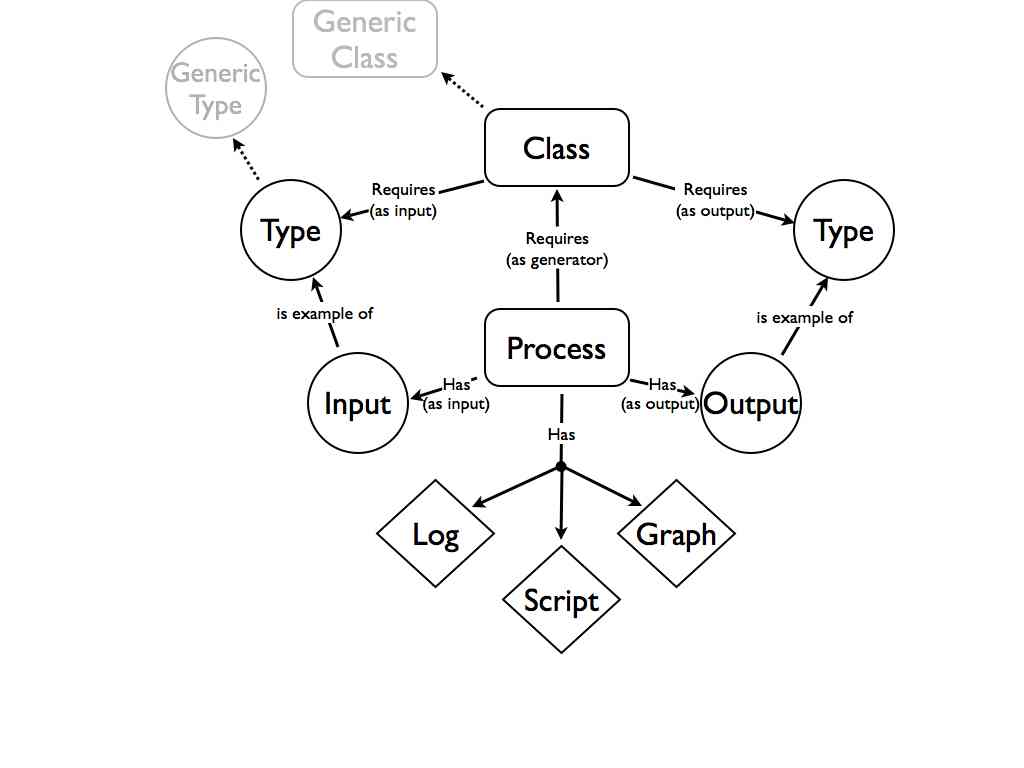
This kind of approach turns out to work quite well in the lab. In our blog based notebook we use a one item-one post approach where every research artifact gets its own URL. Both the verbs, the procedures, and the nouns, the data and materials, all have a unique identifier. The relationships between verbs and nouns is provided by simple links. Thus the structured vocabulary of the lab notebook is [Material] was input to [Process] which generated [Data] (where Material and Data can be interchanged depending on the process). This is not so much 80/20 as 30/70 but even in this very basic form in can be quite useful. Along with records of who did something and when, and some basic tagging this actually makes a quite an effective lab notebook system.
The question is, how can we move beyond this to create a record which is richer enough to provide a real step up, but doesn’t bother the user any more than is necessary and justified by the extra functionality that they’re getting. In fact, ideally we’d capture a richer and more useful record while bothering the user less. A part of the solution lies in the work that Jeremy Frey’s group have done with blogging instruments. By having an instrument create a record of it’s state, inputs and outputs, the user is freed to focus on what their doing, and only needs to link into that record when they start to do their analysis.
Another route is the approach that Peter Murray-Rust’s group are exploring with interactive lab equipment, particularly a fume cupboard that can record spoken instructions and comments and track where objects are, monitoring an entire process in detail. The challenge in this approach lies in translating that information into something that is easy to use downstream. Audio and video remain difficult to search and worth with. Speech recognition isn’t great for formatting and clear presentation.
In the spirit of a limited vocabulary another approach is to use a lightweight infrastructure to record short comments, either structured, or free text. A bakery in London has a switch on its wall which can be turned to one of a small number of baked good as a batch goes into the oven. This is connected to a very basic twitter client then tells the world that there are fresh baked baguettes coming in about twenty minutes. Because this output data is structured it would in principle be possible to track the different baking times and preferences for muffins vs doughnuts over the day and over the year.
The lab is slightly more complex than a bakery. Different processes would take different inputs. Our hypothetical structured vocabulary would need to enable the construction of sentences with subjects, predicates, and objects, but as we’ve learnt with the lab notebook, even the simple predicate “is input toâ€, “is output of†can be very useful. “I am doing X” where X is one of a relatively small set of options provides real time bounds on when important events happened. A little more sophistication could go a long way. A very simple twitter client that provided a relatively small range of structured statements could be very useful. These statements could be processed downstream into a more directly useable record.
Last week I recorded the steps that I carried out in the lab via the hashtag #tweetthelab. These free text tweets make a serviceable, if not perfect, record of the days work. What is missing is a URI for each sample and output data file, and links between the inputs, the processes, and the outputs. But this wouldn’t be too hard to generate, particularly if instruments themselves were actually blogging or tweeting its outputs. A simple client on a tablet, phone, or locally placed computer would make it easy to both capture and to structure the lab record. There is still a need for free text comments and any structured description will not be able to capture everything but the potential for capturing a lot of the detail of what is happening in a lab, as it happens, is significant. And it’s the detail that often isn’t recorded terribly well, the little bits and pieces of exactly when something was done, what did the balance really read, which particular bottle of chemical was picked up.
Twitter is often derided as trivial, as lowering the barrier to shouting banal fragments to the world, but in the lab we need tools that will help us collect, aggregate and structure exactly those banal pieces so that we have them when we need them. Add a little bit of structure to that, but not too much, and we could have a winner. Starting from human discourse always seemed too hard for me, but starting with identifying the simplest things we can say that are also useful to the scientist on the ground seems like a viable route forward.