As I mentioned a couple of weeks or so ago I’ve been playing around with Friendfeed. This is a ‘lifestreaming’ web service which allows you to aggregate ‘all’ of the content you are generating on the web into one place (see here for mine). This is interesting from my perspective because it maps well onto our ideas about generating multiple data streams from a research lab. This raw data then needs to be pulled together and turned into some sort of narrative description of what happened. Continue reading “Friendfeed, lifestreaming, and workstreaming”
The structured experiment
More on the discussion of structured vs unstructured experiment descriptions. Frank has put up a description of the Minimal Information about a Neuroscience Investigation standard at Nature Precedings which comes out of the CARMEN project. Neil Saunder’s has also made some comments on the resistance amongst the lab monkeys to think about structure. Lots of good points here. I wanted to pick out a couple in particular;
From Neil;
My take on the problem is that biologists spend a lot of time generating, analysing and presenting data, but they don’t spend much time thinking about the nature of their data. When people bring me data for analysis I ask questions such as: what kind of data is this? ASCII text? Binary images? Is it delimited? Can we use primary keys? Not surprisingly this is usually met with blank stares, followed by “well…I ran a gel…â€.
Part of this is a language issue. Computer scientists and biologists actually mean something quite different when they refer to ‘data’. For a comp sci person data implies structure. For a biologist data is something that requires structure to be made comprehensible. So don’t ask ‘what kind of data is this?’, ask ‘what kind of file are you generating?’. Most people don’t even know what a primary key is, including me as demonstrated by my misuse of the term when talking about CAS numbers which lead to significant confusion.
I do believe that any experiment [CN – my emphasis] can be described in a structured fashion, if researchers can be convinced to think generically about their work, rather than about the specifics of their own experiments. All experiments share common features such as: (1) a date/time when they were performed; (2) an aim (â€generate PCR productâ€, “run crystal screen for protein Xâ€); (3) the use of protocols and instruments; (4) a result (correct size band on a gel, crystals in well plate A2). The only free-form part is the interpretation.
Here I disagree, but only at the level of detail. The results of any experiment can probably be structured after the event. But not all experiments can be clearly structured either in advance, or as they happen. Many can, and here Neil’s point is a good one, by making some slight changes in the way people think about their experiment much more structure can be captured. I have said before that the process of using our ‘unstructured’ lab book system has made me think and plan my experiments more carefully. Nonetheless I still frequently go off piste, things happen. What started as an SDS-PAGE gel turns into something else (say a quick column on the FPLC).
Without wishing to pick a fight, most people with a computer science background who lean towards the heavily semantic end of the spectrum are dealing with the wet lab scientists after the data has been taken and partially processed. I don’t disagree that it would help the comp sci people if the experimenters worked harder at structuring the data as they generate it, and I do think in general this is a good thing. The problem is that it doesn’t map well onto how the work is actually carried out. The solution I think is a mixture of the free form approach combined with useful tools and widgets that do two things: firstly they make the process of capturing the process easier; secondly the encourage the collection and structuring of data as it comes off. This is what the templates in our system do, and there is no reason in principle why they couldn’t be driven by agreed data models.
Actually the Frey group (who have done the development of the LaBLog system) already have a highly semantic lab book system developed during the MyTea project. One of our future aims is to take the best of both forward into a ‘semi-semantic’ or ‘freely semantic’ system. One of the main problems with implementing the MyTea notebook is that it requires data models. It was developed for synthetic chemistry but it would make sense, in expanding it into the biochemistry/molecular biology area to utilise existing data models with FuGE the obvious main source.
One more point: we need to teach students that every activity leading to a result is an experiment. From my time as a Ph.D. student in the wet lab, I remember feeling as though my day-to-day activities: PCR reactions, purifications, cloning weren’t really experiments […] Experiments were clever, one-shot procedures performed by brilliant postdocs to answer big questions […] Break your activities into steps and ways to describe them as structured data should suggest themselves.
This is very true, and harks back to my comment about language. A lot of the issues here are actually because we mean very different things by ‘experiment’. We probably should use better words, although I think procedure and protocol are similarly loaded with conflicting meanings. Control of language is important and agreement on meaning is, after all, at the root of semantics (or is that semiotics, I’m never sure…)
Incorporating My Experiment and Taverna into the LaBLog – A possible example
During the workshop in late February we had discussions about possible implementations of Taverna work flows to automate specific processes to make our life easier. One specific example we discussed was the reduction and initial analysis of Small Angle Neutrons Scattering data. Here I want to describe a bit of the background to what this is and what we might do to kick of the discussion. Continue reading “Incorporating My Experiment and Taverna into the LaBLog – A possible example”
Proposing a data model for Open Notebooks
‘No data model survives contact with reality’ – Me, Cosener’s House Workshop 29 February 2008
This flippant comment was in response to (I think) Paolo Missier asking me ‘what the data model is’ for our experiments. We were talking about how we might automate various parts of the blog system but the point I was making was that we can’t have a data model with any degree of specificity because we very quickly find the situation where they don’t fit. However, having spent some time thinking about machine readability and the possibility of converting a set of LaBLog posts to RDF, as well as the issues raised by the problems we have with tables, I think we do need some sort of data model. These are my initial thoughts on what that might look like. Continue reading “Proposing a data model for Open Notebooks”
A (small) Feeding Frenzy
Following on from (but unrelated to) my post last week about feed tools we have two posts, one from Deepak Singh, and one from Neil Saunders, both talking about ‘friend feeds’ or ‘lifestreams’. The idea here is of aggregating all the content you are generating (or is being generated about you?) into one place. There are a couple of these about but the main ones seem to be Friendfeed and Profiliac. See Deepaks’s post (or indeed his Friendfeed) for details of the conversations that can come out of these type of things.
What piqued my interest though was the comment Neil made at the bottom of his post about Workstreams.
Here’s a crazy idea – the workstream:
* Neil parsed SwissProt entry Q38897 using parser script swiss2features.pl
* Bob calculated all intersubunit contacts in PDB entry 2jdq using CCP4 package contact
This is exactly the kind of thing I was thinking about as the raw material for the aggregators that would suggest things that you ought to look at, whether it be a paper, a blog post, a person, or a specific experimental result. This type of system will rely absolutely on the willingness of people to make public what they are reading, doing, even perhaps thinking. Indeed I think this is the raw information that will make another one of Neil’s great suggestions feasible.
Following on from Neil’s post I had a short conversation with Alf in the comments about blogging (or Twittering) machines. Alf pointed out a really quite cool example. This is something that we are close to implementing in the open in the lab at RAL. We hope to have the autoclave, PCR machine, and balances all blogging out what they are seeing. This will generate a data feed that we can use to pull specific data items down into the LaBLog.
Perhaps more interesting is the idea of connecting this to people. At the moment the model is that the instruments are doing the blogging. This is probably a good way to go because it keeps a straightforward identifiable data stream. At the moment the trigger for the instruments to blog is a button. However at RAL we use RFID proximity cards for access to the buildings. This means we have an easy identifier for people, so what we aim to do is use the RFID card to trigger data collection (or data feeding).
If this could be captured and processed there is the potential for capturing a lot of the detail of what has happened in the laboratory. Combine this with a couple of Twitter posts giving a little more personal context and it may be possible to reconstruct a pretty complete record of what was done and precisely when. The primary benefit of this would be in trouble shooting but if we could get a little bit of processing into this, and if there are specific actions with agreed labels, then it may be possible to automatically create a large portion of the lab book record.
This may be a great way of recording the kind of machine readable description of experiments that Jean-Claude has been posting about. Imagine a simplistic Twitter interface where you have a limited set of options (I am stirring, I am mixing, I am vortexing, I have run a TLC, I have added some compound). Combine this with a balance, a scanner, and a heating mantle which are blogging out what they are currently seeing, and a barcode reader (and printer) so as to identify what is being manipulated and which compound is which.
One of the problems we have with our lab books is that they can never be detailed enough to capture everything that somebody might be interested in one day. However at the same time they are too detailed for easy reading by third parties. I think there is general agreement that on top of the lab book you need an interpretation layer, an extra blog that explains what is going on to the general public. Perhaps by capturing all the detailed bits automatically we can focus on planning and thinking about the experiments rather than worrying about how to capture everything manually. Then anyone can mash up the results, or the discussion, or the average speed of the stirrer bar, any way they like.
Who’s got the bottle?
Lots of helpful comments from people on my question about what to use as a good identifier of chemicals? I thought it might be useful to re-phrase what it was that I wanted because I think some of the comments, while important discussion points don’t really impinge directly on my current issue.
I have in mind a special type of page on our LaBLog system that will easily allow the generation of a post that describes a new bottle of material that comes into the lab. From a user perspective you want to enter the minimum amount of necessary information, probably a name, a company, perhaps a lot number and/or catalogue number to enable reordering. From the system perspective you want to try and grab as many different ways of describing the material as possible, including where appropriate SMILES, InChi, CML, or whatever. My question was, how do I provide a simple key that will enable the system to go off and find (if possible) these other identifiers. This isn’t really a database per se but a collection of descriptors on a page (although we would like to pull the data out and into a proper database at a later stage). CAS numbers are great because they are written on most bottles and are a well curated system. However I thought that the only way of converting from CAS to anything else was to go through a CAS service. Therefore I thought PubChem CID’s (or SID’s) might be a good way to do this.
So from my perspective a lot of the technical issues with substances versus chemicals versus structures aren’t so important. All I want is to, on a best efforts basis, pull down as many other descriptors as possible to expose in the post. For some things (e.g. yeast extract) the issues of substances versus compounds (not to mention supplier) get right out of hand (I am slightly bemused that it has a CAS number, and there are multiple SID’s in PubChem). Certainly it ain’t going to have an InChi. But if you try and get nothing it doesn’t really matter. Also we are dealing here with common materials. If as Dan Zaharevitz points out, we were dealing with compounds from synthetic chemists we would get into serious trouble, but in this case I think we could rely on our collaborating chemists to get InChi’s/SMILES/CML correct and use those directly. In the ideal Open Notebook Science world we would simply point to their lab books anyway.
So the fundamental issue for me; is there something written on the bottle of material that we can use as a convenient search key to pull down as many other descriptors as we can?
Now I am with Antony Williams on this, if CAS got its act together and made their numbers an open standard then that would be the best solution. It is curated and all pervasive as an identifier. Both Antony and Rich Apodaca have pointed out that I was wrong to say that CAS numbers aren’t in PubChem (and Rich pointed to two useful posts [1], [2] on how to get into PubChem using CAS numbers). So actually, my problem is probably solved by an application of Rich’s instructions on hacking PubChem (even if it turns out we have to download the entire database). The issue here is whether they will stay there or whether they may in the end get pulled.
I do think that for my purposes that PubChem CID’s and SID’s will do the job in this specific case. However as has been pointed out there are issues with reliability and curation. So I will accept that it is probably too early to start suggesting that suppliers label their bottles with PubChem IDs. This may happen anyway (Aldrich seem to have them in the catalogue at least; haven’t been able to check a bottle yet) in the longer term and I guess we have to wait and see what happens.
Peter Murray-Rust has also updated with a series of posts [1], [2], [3] around the issues of chemical substance identity, CAS, Wikipedia et al. Peter Suber has aggregated many of the related posts together. And Glyn Moody has called us to the barricades.
Give me the feed tools and I can rule the world!
Two things last week gave me more cause to think a bit harder about the RSS feeds from our LaBLog and how we can use them. First, when I gave my talk at UKOLN I made a throwaway comment about search and aggregation. I was arguing that the real benefits of open practice would come when we can use other people’s filters and aggregation tools to easily access the science that we ought to be seeing. Google searching for a specific thing isn’t enough. We need to have an aggregated feed of the science we want or need to see delivered automatically. i.e. we need systems to know what to look for even before the humans know it exists. I suggested the following as an initial target;
‘If I can automatically identify all the compounds recently made in Jean-Claude’s group and then see if anyone has used those compounds [or similar compounds] in inhibitor screens for drug targets then we will be on our way towards managing the information’
The idea here would be to take a ‘Molecules’ feed (such as the molecules Blog at UsefulChem or molecules at Chemical Blogspace) extract the chemical identifiers (InChi, Smiles, CML or whatever) and then use these to search feeds from those people exposing experimental results from drug screening. You might think some sort of combination of Yahoo! Pipes and Google Search ought to do it.
So I thought I’d give this a go. And I fell at the first hurdle. I could grab the feed from the UsefulChem molecules Blog but what I actually did was set up a test post in the Chemtools Sandpit Blog. Here I put the InChi of one of the compounds from UsefulChem that was recently tested as a falcipain 2 inhibitor. The InChi went in as both clear text and as the microformat approach suggested by Egon Willighagen. Pipes was perfectly capable of pulling the feed down, and reducing it to only the posts that contained InChi’s but I couldn’t for the life of me figure out how to extract the InChi itself. Pipes doesn’t seem to see microformats. Another problem is that there is no obvious way of converting a Google Search (or Google Custom Search) to an RSS feed.
Now there may well be ways to do this, or perhaps other tools to do it better but they aren’t immediately obvious to me. Would the availability of such tools help us to take the Open Research agenda forwards? Yes, definitely. I am not sure exactly how much or how fast but without easy to use tools, that are well presented, and easily available, the case for making the information available is harder to make. What’s the point of having it on the cloud if you can’t customise your aggregation of it? To me this is the killer app; being able to identify, triage, and collate data as it happens with easily useable and automated tools. I want to see the stuff I need to see in feed reader before I know it exists. Its not that far away but we ain’t there yet.
The other thing this brought home to me was the importance of feeds and in particular of rich feeds. One of the problems with Wikis is that they don’t in general provide an aggregated or user configurable feed of the site in general or a name space such as a single lab book. They also don’t readily provide a means of tagging or adding metadata. Neither Wikis nor Blogs provide immediately accessible tools that provide the ability to configure multiple RSS feeds, at least not in the world of freely hosted systems. The Chemtools blogs each put out an RSS feed but it doesn’t currently include all the metadata. The more I think about this the more crucial I think it is.
To see why I will use another example. One of the features that people liked about our Blog based framework at the workshop last week was the idea that they got a catalogue of various different items (chemicals, oligonucleotides, compound types) for free once the information was in the system and properly tagged. Now this is true but you don’t get the full benefits of a database for searching, organisation, presentation etc. We have been using DabbleDB to handle a database of lab materials and one of our future goals has been to automatically update the database. What I hadn’t realised before last week was the potential to use user configured RSS feeds to set up multiple databases within DabbleDB to provide more sophisticated laboratory stocks database.
DabbleDB can be set up to read RSS or other XML or JSON feeds to update as was pointed out to me by Lucy Powers at the workshop. To update a database all we need is a properly configured RSS feed. As long as our templates are stable the rest of the process is reasonably straightforward and we can generate databases of materials of all sorts along with expiry dates, lot numbers, ID numbers, safety data etc etc. The key to this is rich feeds that carry as much information as possible, and in particular as much of the information we have chosen to structure as possible. We don’t even need the feeds to be user configurable within the system itself as we can use Pipes to easily configure custom feeds.
We, or rather a noob like me, can do an awful lot with some of the tools already available and a bit of judicious pointing and clicking. When these systems are just a little bit better at extracting information (and when we get just a little bit better at putting information in, by making it part of the process) we are going to be doing lots of very exciting things. I am trying to keep my diary clear for the next couple of months…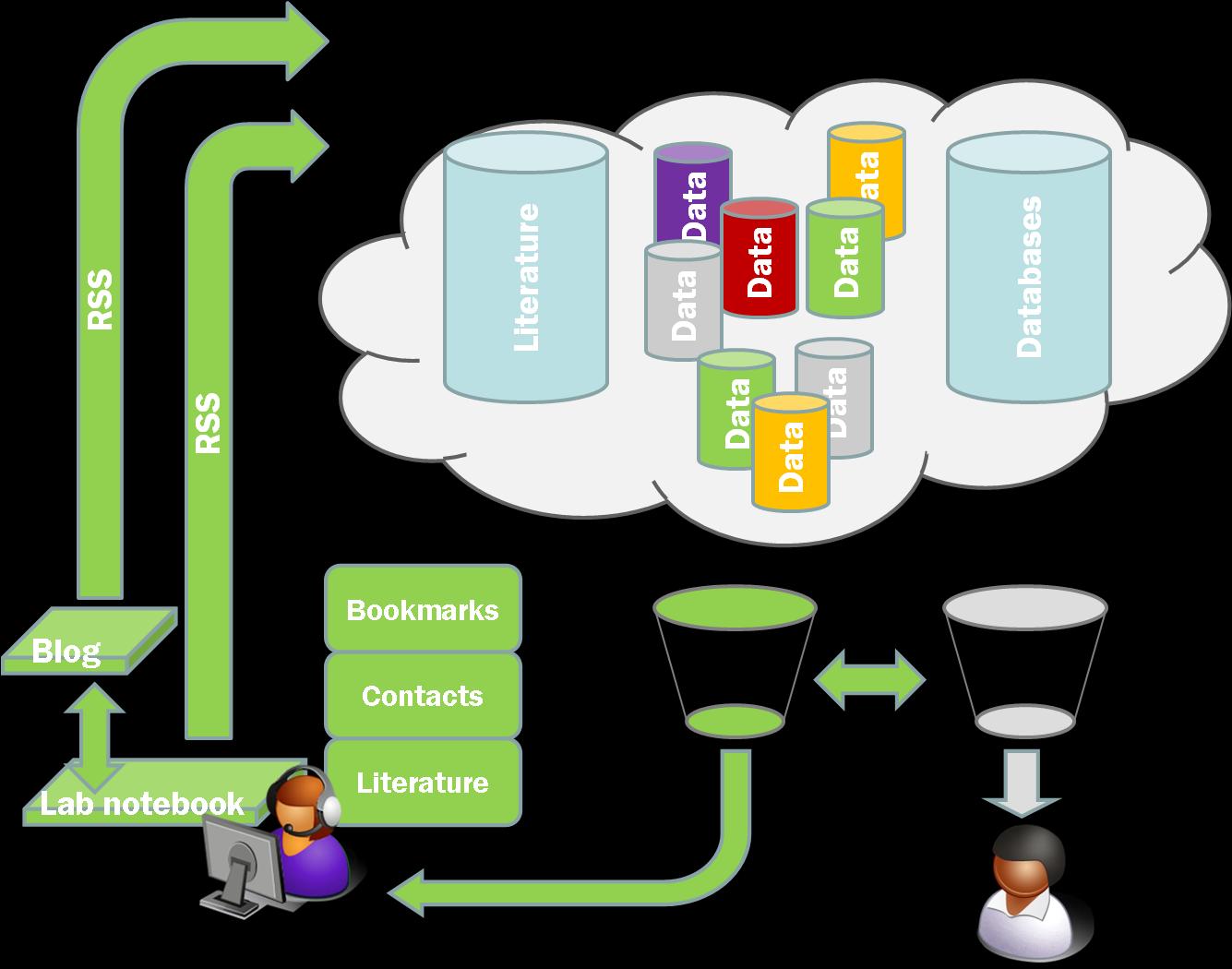
Workshop on Blog Based Notebooks
DUE TO SEVERE COMMENT SPAM ON THIS POST I HAVE CLOSED IT TO COMMENTS
On February 28/29 we held a workshop on our Blog Based notebook system at the Cosener’s House in Abingdon, Oxfordshire. This was a small workshop with 13 people including biochemists (from Southampton, Cardiff, and RAL), social scientists (from Oxford Internet Institute and Computing Laboratory), developers from the MyGrid and MyExperiment family and members of the the Blog development team. The purpose of the workshop was to try and identify both the key problems that need to be addressed in the next version of the Blog Based notebook system and also to ask the question ‘Does this system deliver functionality that people want’. Another aim was to identify specific ways in which we could interact with MyExperiment to deliver enhanced functionality for us as well as new members for MyExperiment.
