
- Image by cameronneylon via Flickr
I wrote a few weeks back about the idea of re-imagining the formally published scientific paper as an aggregation of objects. The idea behind this is that it provides both continuity, through enabling the display of such papers in more or less the same way as we do currently, enhancing functionality by, for instance, embedding active versions of figures in a native form, and at the same time providing a route towards a linked data web for research.
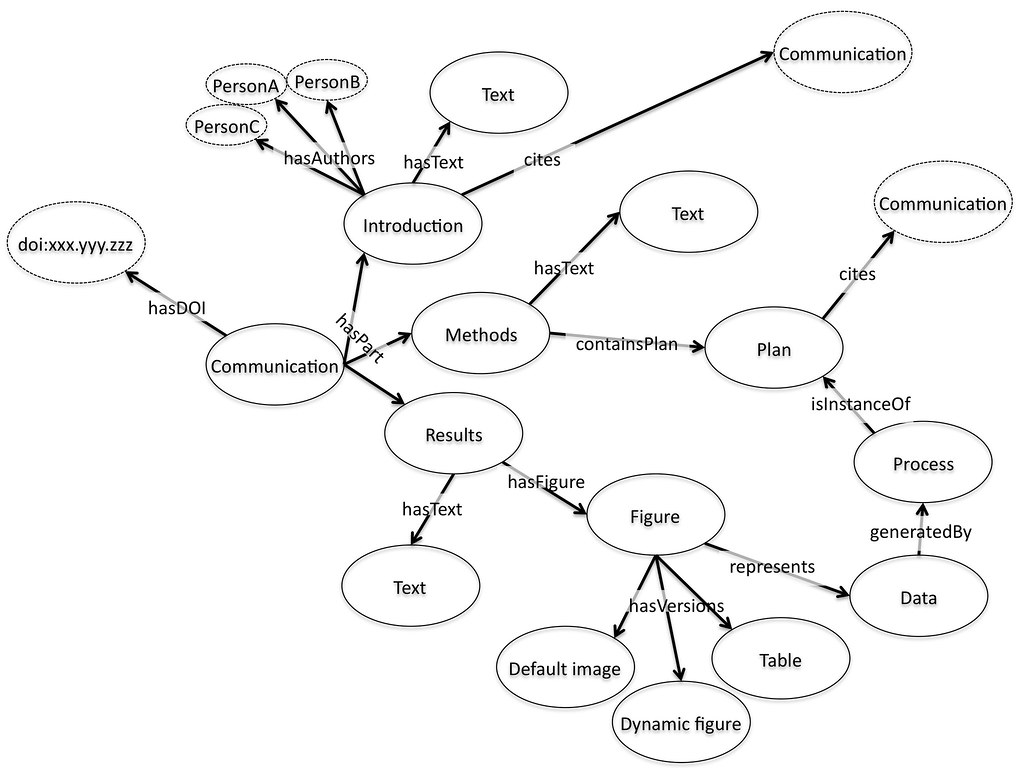
Fundamentally the idea is that we publish fragments, and then aggregate these fragments together. The mechanism of aggregation is supposed to be an expanded version of the familiar paradigm of citation: the introduction of a paper will link to and cite other papers as usual, and these would be incorporated as links and citations within the presentation of the paper. But in addition the paper will cite the introduction. By default the introduction would be included in the view of the paper presented to the user, but equally the user might choose to only look at figures, only conclusions, or only the citations. Figures would be citations to data, again available on the web, again with a default visualization that might be an embedded active graph or a static image.
I asserted that the tools for achieving this are more or less in place. Actually that is only half true. The tools for storing, displaying, and even to some extent archiving communications in this form do exist, at least in the form of examples.
An emerging standard for aggregated objects on the web is the Open Archives Initiative – Object Re-use and Exchange (OAI-ORE). The OAI-ORE object is a generic description of the address of a series of things on the web and how they relate to each other. It is the natural approach for representing the idea of a paper as an aggregation of pieces. The OAI-ORE object itself is RDF with no concept of how it should be displayed or used. It is just a set of things, each labeled with their role in the overall object. In principle at least makes it straightforward to display it in any number of ways. A simple example would be converting OAI-ORE to the NLM-DTD XML format. The devil as always, is in the detail here but this makes a good first pass technical requirement for the details of how the pieces of the OAI-ORE object are described; it must be straightforward to convert to NLM-DTD.
Once we have both the collection of pieces, their relationship to each other, and the idea that we can choose to display some or all of these pieces in any way we choose then a lot of the rest falls into place. Figures can be data objects, which have a default visualization method. These visualizations can be embedded in the way which is now familiar with audio and video files. But equally references to gene names, structures, and chemical entities could be treated the same way. Want the chemical name? Just click a button, the visualization tool will deliver that. Want the structure? Again, just click the button, toggle the menu, or write the script to ask for it in that form if you are doing that kind of thing. We would need more open standards for embedding objects; probably less Flash, and more open standards but that’s a fairly minor issue.
There needs to be some communication between the citing object (the paper) and the cited object (the data, figure, text, external reference). This could be built up from the TrackBack or Pingback protocols. There also needs to be default content negotiation: “I want this data, what can you give me? Graph? Table?…ok I’ll take the graph…†That’s just a RESTful API, something which is more or less standard for the consumer web data services but which is badly missing on the research web. None of this is actually terribly difficult and there are good tools out there to do it.
But I said that we only had half solved. The other side of the problem is good authoring and publishing tools. All of the above assumes that these OAI-ORE objects exist or can be easily built, and that the pieces we want to aggregate are already on the web, ready to be pointed at and embedded. They are not. We have two fundamental problems. First we have to get these things onto the web in a useful and re-useable form. Some of this can be done with existing data services such as ChemSpider, Genbank, PubChem, GEO, etc. but that is the easy end of the problem. The hard bit is the heterogenous mass of pieces of data, Excel spreadsheets, CSV files, XML and binaries, that make up the majority of the research outputs we generate.
Publication could be made easy, using automatic upload tools and lightweight data services that provide a place for them on the web. The criticism is often made that “just publishing†is not enough because there is no context. What is often missed is that the best way to provide context is for the person who generated the research object to link it in to a larger record. The catch is, for that to be useful they have to publish it to the web first, otherwise the record they create points at a local and inaccessible object. So we need tools that simply push the raw material up onto the web, probably in the short to medium term to secure servers, but ones where the individual objects can be made public at some point.
So the other tools we need are for authoring these documents. These will look and behave like a Word Processor (or like a LaTeX document for those who prefer that route) but with a clever reference manager and citation creator. Today our reference libraries only contain papers. But imagine that your library contained all of the data you’ve generated as well and that the easiest way to write up your lab notebook was to simply right click and include the reference, select the visualization that you want and you’re done. All the details of default visualizations, of where the data really is, of adding the record to the OAI-ORE root node, all of this is done for you behind the scenes. You might need to select lumps of text to say what their role is, probably using some analogue of styles, similar to the way that the Integrated Content Environment (ICE) system does.
This environment could be built for Word, for Open Office, for LaTeX. One of the reasons I remain excited about Google Wave is that it should be relatively easy to prototype such an environment, the hooks are already there in a way that they aren’t in traditional document authoring tools because Wave is much more web native. It will however take quite a lot of work. There is also a chicken and egg problem in that such an environment isn’t a whole lot of use without the published objects to aggregate together, and the publication services to provide rich views of the final aggregated documents. It will be quite a lot of work to build all of these pieces, and it will take some time before the benefits become clear. But I think it is a direction that is worth pursuing because it takes the best of what we already know works on the web and applies it to an evolutionary adaption of the communication style that is already familiar. The revolution comes once the pieces are there for people to work with in new ways.
Related articles by Zemanta
- Sefton, Peter: ICE and word processor / HTML interop, the ugly, uglier, ugliest (ptsefton.com)
- Main Articles: ‘Enhancing Scientific Communication through Aggregated Publications Environments’, Ariadne Issue 61 (ariadne.ac.uk)
- Bigwood, David: Exposing OAI Content (catalogablog.blogspot.com)

![Reblog this post [with Zemanta]](http://img.zemanta.com/reblog_e.png?x-id=219f7020-8ab0-4a39-b877-01f47779fc1b)
![Reblog this post [with Zemanta]](http://img.zemanta.com/reblog_e.png?x-id=8af89e7c-59ca-438d-8e68-4efa40cffd79)