“In the future everyone will be a journal editor for 15 minutes” – apologies to Andy Warhol
Suddenly it seems everyone wants to re-imagine scientific communication. From the ACS symposium a few weeks back to a PLoS Forum, via interesting conversations with a range of publishers, funders and scientists, it seems a lot of people are thinking much more seriously about how to make scientific communication more effective, more appropriate to the 21st century and above all, to take more advantage of the power of the web.
For me, the “paper” of the future has to encompass much more than just the narrative descriptions of processed results we have today. It needs to support a much more diverse range of publication types, data, software, processes, protocols, and ideas, as well provide a rich and interactive means of diving into the detail where the user in interested and skimming over the surface where they are not. It needs to provide re-visualisation and streaming under the users control and crucially it needs to provide the ability to repackage the content for new purposes; education, public engagement, even main stream media reporting.
I’ve got a lot of mileage recently out of thinking about how to organise data and records by ignoring the actual formats and thinking more about what the objects I’m dealing with are, what they represent, and what I want to do with them. So what do we get if we apply this thinking to the scholarly published article?
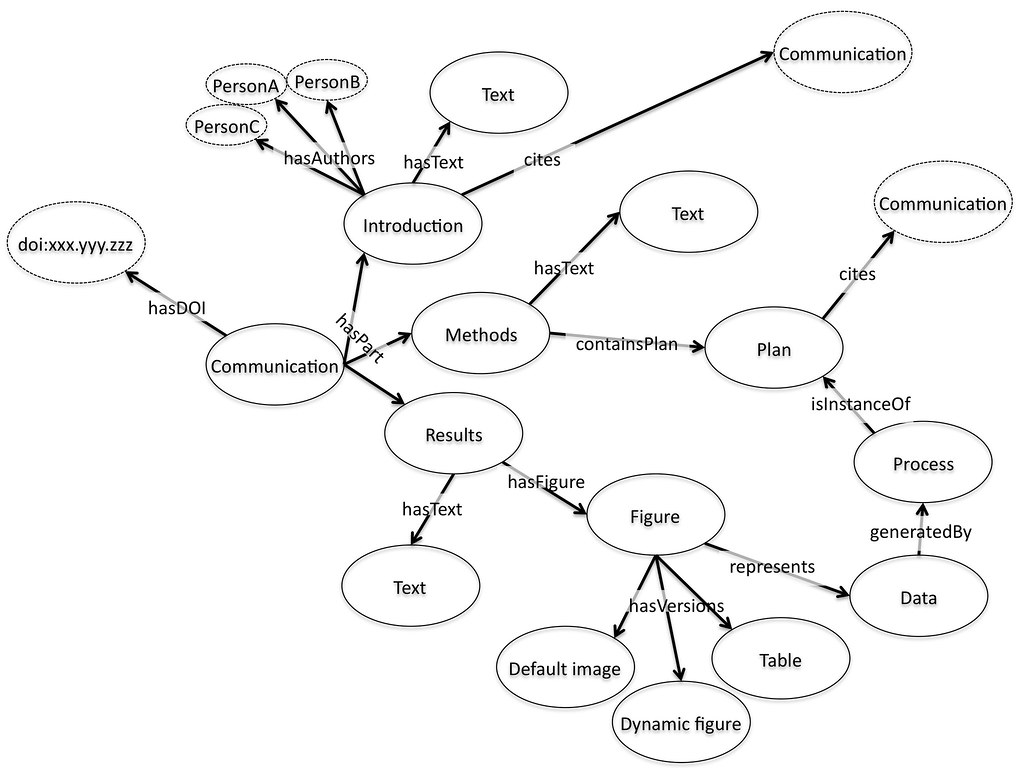
For me, a paper is an aggregation of objects. It contains, text, divided up into sections, often with references to other pieces of work. Some of these references are internal, to figures and tables, which are representations of data in some form or another. The paper world of journals has led us to think about these as images but a much better mental model for figures on the web is of an embedded object, perhaps a visualisation from a service like Many Eyes, Swivel, and Tableau Public. Why is this better? It is better because it maps more effectively onto what we want to do with the figure. We want to use it to absorb the data it represents, and to do this we might want to zoom, pan, re-colour, or re-draw the data. But we want to know if we do this that we are using the same underlying data, so the data needs a home, an address somewhere on the web, perhaps with the journal, or perhaps somewhere else entirely, that we can refer to with confidence.
If that data has an individual identity it can in turn refer back to the process used to generate it, perhaps in an online notebook or record, perhaps pointing to a workflow or software process based on another website. Maybe when I read the paper I want that included, maybe when you read it you don’t – it is a personal choice, but one that should be easy to make. Indeed, it is a choice that would be easy to make with today’s flexible web frameworks if the underlying pieces were available and represented in the right way.
The authors of the paper can also be included as a reference to a unique identifier. Perhaps the authors of the different segments are different. This is no problem, each piece can refer to the people that generated it. Funders and other supporting players might be included by reference. Again this solves a real problem of today, different players are interested in how people contributed to a piece of work, not just who wrote the paper. Providing a reference to a person where the link show what their contribution was can provide this much more detailed information. Finally the overall aggregation of pieces that is brought together and finally published also has a unique identifier, often in the form of the familiar DOI.
This view of the paper is interesting to me for two reasons. The first is that it natively supports a wide range of publication or communication types, including data papers, process papers, protocols, ideas and proposals. If we think of publication as the act of bringing a set of things together and providing them with a coherent identity then that publication can be many things with many possible uses. In a sense this is doing what a traditional paper should do, bringing all the relevant information into a single set of pages that can be found together, as opposed to what they usually do, tick a set of boxes about what a paper is supposed to look like. “Is this publishable?” is an almost meaningless question on the web. Of course it is. “Is it a paper?” is the question we are actually asking. By applying the principles of what the paper should be doing as opposed to the straightjacket of a paginated, print-based document, we get much more flexibility.
The second aspect which I find exciting revolves around the idea of citation as both internal and external references about the relationships between these individual objects. If the whole aggregation has an address on the web via a doi or a URL, and if its relationship both to the objects that make it up and to other available things on the web are made clear in a machine readable citation then we have the beginnings of a machine readable scientific web of knowledge. If we take this view of objects and aggregates that cite each other, and we provide details of what the citations mean (this was used in that, this process created that output, this paper is cited as an input to that one) then we are building the semantic web as a byproduct of what we want to do anyway. Instead of scaring people with angle brackets we are using a paradigm that researchers understand and respect, citation, to build up meaningful links between packages of knowledge. We need the authoring tools that help us build and aggregate these objects together and tools that make forming these citations easy and natural by using the existing ideas around linking and referencing but if we can build those we get the semantic web for science as a free side product – while also making it easier for humans to find the details they’re looking for.
Finally this view blows apart the monolithic role of the publisher and creates an implicit marketplace where anybody can offer aggregations that they have created to potential customers. This might range from a high school student putting their science library project on the web through to a large scale commercial publisher that provides a strong brand identity, quality filtering, and added value through their infrastructure or services. And everything in between. It would mean that large scale publishers would have to compete directly with the small scale on a value-for-money basis and that new types of communication could be rapidly prototyped and deployed.
There are a whole series of technical questions wrapped up in this view, in particular if we are aggregating things that are on the web, how did they get there in the first place, and what authoring tools will we need to pull them together. I’ll try to start on that in a follow-up post.

![Reblog this post [with Zemanta]](http://img.zemanta.com/reblog_e.png?x-id=8af89e7c-59ca-438d-8e68-4efa40cffd79)